Doc modernization (#3474)
* Change doc build to reST exclusively * Rewrite Intro doc in reST; create toctree * Update parameter and contribute * Convert tutorials to reST * Convert Python tutorials to reST * Convert CLI and Julia docs to reST * Enable markdown for R vignettes * Done migrating to reST * Add guzzle_sphinx_theme to requirements * Add breathe to requirements * Fix search bar * Add link to user forum
This commit is contained in:
parent
c004cea788
commit
05b089405d
@ -1,17 +0,0 @@
|
||||
XGBoost R Package
|
||||
=================
|
||||
[](http://cran.r-project.org/web/packages/xgboost)
|
||||
[](http://cran.rstudio.com/web/packages/xgboost/index.html)
|
||||
|
||||
|
||||
You have found the XGBoost R Package!
|
||||
|
||||
Get Started
|
||||
-----------
|
||||
* Checkout the [Installation Guide](../build.md) contains instructions to install xgboost, and [Tutorials](#tutorials) for examples on how to use xgboost for various tasks.
|
||||
* Please visit [walk through example](../../R-package/demo).
|
||||
|
||||
Tutorials
|
||||
---------
|
||||
- [Introduction to XGBoost in R](xgboostPresentation.md)
|
||||
- [Discover your data with XGBoost in R](discoverYourData.md)
|
||||
28
doc/R-package/index.rst
Normal file
28
doc/R-package/index.rst
Normal file
@ -0,0 +1,28 @@
|
||||
#################
|
||||
XGBoost R Package
|
||||
#################
|
||||
|
||||
.. raw:: html
|
||||
|
||||
<a href="http://cran.r-project.org/web/packages/xgboost"><img alt="CRAN Status Badge" src="http://www.r-pkg.org/badges/version/xgboost"></a>
|
||||
<a href="http://cran.rstudio.com/web/packages/xgboost/index.html"><img alt="CRAN Downloads" src="http://cranlogs.r-pkg.org/badges/xgboost"></a>
|
||||
|
||||
You have found the XGBoost R Package!
|
||||
|
||||
***********
|
||||
Get Started
|
||||
***********
|
||||
* Checkout the :doc:`Installation Guide </build>` contains instructions to install xgboost, and :doc:`Tutorials </tutorials/index>` for examples on how to use XGBoost for various tasks.
|
||||
* Read the `API documentation <https://cran.r-project.org/web/packages/xgboost/xgboost.pdf>`_.
|
||||
* Please visit `Walk-through Examples <https://github.com/dmlc/xgboost/tree/master/R-package/demo>`_.
|
||||
|
||||
*********
|
||||
Tutorials
|
||||
*********
|
||||
|
||||
.. toctree::
|
||||
:maxdepth: 2
|
||||
:titlesonly:
|
||||
|
||||
Introduction to XGBoost in R <xgboostPresentation>
|
||||
Understanding your dataset with XGBoost <discoverYourData>
|
||||
23
doc/_static/custom.css
vendored
Normal file
23
doc/_static/custom.css
vendored
Normal file
@ -0,0 +1,23 @@
|
||||
div.breathe-sectiondef.container {
|
||||
width: 100%;
|
||||
}
|
||||
|
||||
div.literal-block-wrapper.container {
|
||||
width: 100%;
|
||||
}
|
||||
|
||||
.red {
|
||||
color: red;
|
||||
}
|
||||
|
||||
table {
|
||||
border: 0;
|
||||
}
|
||||
|
||||
td, th {
|
||||
padding: 1px 8px 1px 5px;
|
||||
border-top: 0;
|
||||
border-bottom: 1px solid #aaa;
|
||||
border-left: 0;
|
||||
border-right: 0;
|
||||
}
|
||||
752
doc/_static/searchtools-new.js
vendored
752
doc/_static/searchtools-new.js
vendored
@ -1,752 +0,0 @@
|
||||
/*
|
||||
* searchtools.js_t
|
||||
* ~~~~~~~~~~~~~~~~
|
||||
*
|
||||
* Sphinx JavaScript utilities for the full-text search.
|
||||
*
|
||||
* :copyright: Copyright 2007-2016 by the Sphinx team, see AUTHORS.
|
||||
* :license: BSD, see LICENSE for details.
|
||||
*
|
||||
*/
|
||||
|
||||
|
||||
/* Non-minified version JS is _stemmer.js if file is provided */
|
||||
/**
|
||||
* Porter Stemmer
|
||||
*/
|
||||
var Stemmer = function() {
|
||||
|
||||
var step2list = {
|
||||
ational: 'ate',
|
||||
tional: 'tion',
|
||||
enci: 'ence',
|
||||
anci: 'ance',
|
||||
izer: 'ize',
|
||||
bli: 'ble',
|
||||
alli: 'al',
|
||||
entli: 'ent',
|
||||
eli: 'e',
|
||||
ousli: 'ous',
|
||||
ization: 'ize',
|
||||
ation: 'ate',
|
||||
ator: 'ate',
|
||||
alism: 'al',
|
||||
iveness: 'ive',
|
||||
fulness: 'ful',
|
||||
ousness: 'ous',
|
||||
aliti: 'al',
|
||||
iviti: 'ive',
|
||||
biliti: 'ble',
|
||||
logi: 'log'
|
||||
};
|
||||
|
||||
var step3list = {
|
||||
icate: 'ic',
|
||||
ative: '',
|
||||
alize: 'al',
|
||||
iciti: 'ic',
|
||||
ical: 'ic',
|
||||
ful: '',
|
||||
ness: ''
|
||||
};
|
||||
|
||||
var c = "[^aeiou]"; // consonant
|
||||
var v = "[aeiouy]"; // vowel
|
||||
var C = c + "[^aeiouy]*"; // consonant sequence
|
||||
var V = v + "[aeiou]*"; // vowel sequence
|
||||
|
||||
var mgr0 = "^(" + C + ")?" + V + C; // [C]VC... is m>0
|
||||
var meq1 = "^(" + C + ")?" + V + C + "(" + V + ")?$"; // [C]VC[V] is m=1
|
||||
var mgr1 = "^(" + C + ")?" + V + C + V + C; // [C]VCVC... is m>1
|
||||
var s_v = "^(" + C + ")?" + v; // vowel in stem
|
||||
|
||||
this.stemWord = function (w) {
|
||||
var stem;
|
||||
var suffix;
|
||||
var firstch;
|
||||
var origword = w;
|
||||
|
||||
if (w.length < 3)
|
||||
return w;
|
||||
|
||||
var re;
|
||||
var re2;
|
||||
var re3;
|
||||
var re4;
|
||||
|
||||
firstch = w.substr(0,1);
|
||||
if (firstch == "y")
|
||||
w = firstch.toUpperCase() + w.substr(1);
|
||||
|
||||
// Step 1a
|
||||
re = /^(.+?)(ss|i)es$/;
|
||||
re2 = /^(.+?)([^s])s$/;
|
||||
|
||||
if (re.test(w))
|
||||
w = w.replace(re,"$1$2");
|
||||
else if (re2.test(w))
|
||||
w = w.replace(re2,"$1$2");
|
||||
|
||||
// Step 1b
|
||||
re = /^(.+?)eed$/;
|
||||
re2 = /^(.+?)(ed|ing)$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
re = new RegExp(mgr0);
|
||||
if (re.test(fp[1])) {
|
||||
re = /.$/;
|
||||
w = w.replace(re,"");
|
||||
}
|
||||
}
|
||||
else if (re2.test(w)) {
|
||||
var fp = re2.exec(w);
|
||||
stem = fp[1];
|
||||
re2 = new RegExp(s_v);
|
||||
if (re2.test(stem)) {
|
||||
w = stem;
|
||||
re2 = /(at|bl|iz)$/;
|
||||
re3 = new RegExp("([^aeiouylsz])\\1$");
|
||||
re4 = new RegExp("^" + C + v + "[^aeiouwxy]$");
|
||||
if (re2.test(w))
|
||||
w = w + "e";
|
||||
else if (re3.test(w)) {
|
||||
re = /.$/;
|
||||
w = w.replace(re,"");
|
||||
}
|
||||
else if (re4.test(w))
|
||||
w = w + "e";
|
||||
}
|
||||
}
|
||||
|
||||
// Step 1c
|
||||
re = /^(.+?)y$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
stem = fp[1];
|
||||
re = new RegExp(s_v);
|
||||
if (re.test(stem))
|
||||
w = stem + "i";
|
||||
}
|
||||
|
||||
// Step 2
|
||||
re = /^(.+?)(ational|tional|enci|anci|izer|bli|alli|entli|eli|ousli|ization|ation|ator|alism|iveness|fulness|ousness|aliti|iviti|biliti|logi)$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
stem = fp[1];
|
||||
suffix = fp[2];
|
||||
re = new RegExp(mgr0);
|
||||
if (re.test(stem))
|
||||
w = stem + step2list[suffix];
|
||||
}
|
||||
|
||||
// Step 3
|
||||
re = /^(.+?)(icate|ative|alize|iciti|ical|ful|ness)$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
stem = fp[1];
|
||||
suffix = fp[2];
|
||||
re = new RegExp(mgr0);
|
||||
if (re.test(stem))
|
||||
w = stem + step3list[suffix];
|
||||
}
|
||||
|
||||
// Step 4
|
||||
re = /^(.+?)(al|ance|ence|er|ic|able|ible|ant|ement|ment|ent|ou|ism|ate|iti|ous|ive|ize)$/;
|
||||
re2 = /^(.+?)(s|t)(ion)$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
stem = fp[1];
|
||||
re = new RegExp(mgr1);
|
||||
if (re.test(stem))
|
||||
w = stem;
|
||||
}
|
||||
else if (re2.test(w)) {
|
||||
var fp = re2.exec(w);
|
||||
stem = fp[1] + fp[2];
|
||||
re2 = new RegExp(mgr1);
|
||||
if (re2.test(stem))
|
||||
w = stem;
|
||||
}
|
||||
|
||||
// Step 5
|
||||
re = /^(.+?)e$/;
|
||||
if (re.test(w)) {
|
||||
var fp = re.exec(w);
|
||||
stem = fp[1];
|
||||
re = new RegExp(mgr1);
|
||||
re2 = new RegExp(meq1);
|
||||
re3 = new RegExp("^" + C + v + "[^aeiouwxy]$");
|
||||
if (re.test(stem) || (re2.test(stem) && !(re3.test(stem))))
|
||||
w = stem;
|
||||
}
|
||||
re = /ll$/;
|
||||
re2 = new RegExp(mgr1);
|
||||
if (re.test(w) && re2.test(w)) {
|
||||
re = /.$/;
|
||||
w = w.replace(re,"");
|
||||
}
|
||||
|
||||
// and turn initial Y back to y
|
||||
if (firstch == "y")
|
||||
w = firstch.toLowerCase() + w.substr(1);
|
||||
return w;
|
||||
}
|
||||
}
|
||||
|
||||
|
||||
|
||||
/**
|
||||
* Simple result scoring code.
|
||||
*/
|
||||
var Scorer = {
|
||||
// Implement the following function to further tweak the score for each result
|
||||
// The function takes a result array [filename, title, anchor, descr, score]
|
||||
// and returns the new score.
|
||||
/*
|
||||
score: function(result) {
|
||||
return result[4];
|
||||
},
|
||||
*/
|
||||
|
||||
// query matches the full name of an object
|
||||
objNameMatch: 11,
|
||||
// or matches in the last dotted part of the object name
|
||||
objPartialMatch: 6,
|
||||
// Additive scores depending on the priority of the object
|
||||
objPrio: {0: 15, // used to be importantResults
|
||||
1: 5, // used to be objectResults
|
||||
2: -5}, // used to be unimportantResults
|
||||
// Used when the priority is not in the mapping.
|
||||
objPrioDefault: 0,
|
||||
|
||||
// query found in title
|
||||
title: 15,
|
||||
// query found in terms
|
||||
term: 5
|
||||
};
|
||||
|
||||
|
||||
|
||||
|
||||
|
||||
var splitChars = (function() {
|
||||
var result = {};
|
||||
var singles = [96, 180, 187, 191, 215, 247, 749, 885, 903, 907, 909, 930, 1014, 1648,
|
||||
1748, 1809, 2416, 2473, 2481, 2526, 2601, 2609, 2612, 2615, 2653, 2702,
|
||||
2706, 2729, 2737, 2740, 2857, 2865, 2868, 2910, 2928, 2948, 2961, 2971,
|
||||
2973, 3085, 3089, 3113, 3124, 3213, 3217, 3241, 3252, 3295, 3341, 3345,
|
||||
3369, 3506, 3516, 3633, 3715, 3721, 3736, 3744, 3748, 3750, 3756, 3761,
|
||||
3781, 3912, 4239, 4347, 4681, 4695, 4697, 4745, 4785, 4799, 4801, 4823,
|
||||
4881, 5760, 5901, 5997, 6313, 7405, 8024, 8026, 8028, 8030, 8117, 8125,
|
||||
8133, 8181, 8468, 8485, 8487, 8489, 8494, 8527, 11311, 11359, 11687, 11695,
|
||||
11703, 11711, 11719, 11727, 11735, 12448, 12539, 43010, 43014, 43019, 43587,
|
||||
43696, 43713, 64286, 64297, 64311, 64317, 64319, 64322, 64325, 65141];
|
||||
var i, j, start, end;
|
||||
for (i = 0; i < singles.length; i++) {
|
||||
result[singles[i]] = true;
|
||||
}
|
||||
var ranges = [[0, 47], [58, 64], [91, 94], [123, 169], [171, 177], [182, 184], [706, 709],
|
||||
[722, 735], [741, 747], [751, 879], [888, 889], [894, 901], [1154, 1161],
|
||||
[1318, 1328], [1367, 1368], [1370, 1376], [1416, 1487], [1515, 1519], [1523, 1568],
|
||||
[1611, 1631], [1642, 1645], [1750, 1764], [1767, 1773], [1789, 1790], [1792, 1807],
|
||||
[1840, 1868], [1958, 1968], [1970, 1983], [2027, 2035], [2038, 2041], [2043, 2047],
|
||||
[2070, 2073], [2075, 2083], [2085, 2087], [2089, 2307], [2362, 2364], [2366, 2383],
|
||||
[2385, 2391], [2402, 2405], [2419, 2424], [2432, 2436], [2445, 2446], [2449, 2450],
|
||||
[2483, 2485], [2490, 2492], [2494, 2509], [2511, 2523], [2530, 2533], [2546, 2547],
|
||||
[2554, 2564], [2571, 2574], [2577, 2578], [2618, 2648], [2655, 2661], [2672, 2673],
|
||||
[2677, 2692], [2746, 2748], [2750, 2767], [2769, 2783], [2786, 2789], [2800, 2820],
|
||||
[2829, 2830], [2833, 2834], [2874, 2876], [2878, 2907], [2914, 2917], [2930, 2946],
|
||||
[2955, 2957], [2966, 2968], [2976, 2978], [2981, 2983], [2987, 2989], [3002, 3023],
|
||||
[3025, 3045], [3059, 3076], [3130, 3132], [3134, 3159], [3162, 3167], [3170, 3173],
|
||||
[3184, 3191], [3199, 3204], [3258, 3260], [3262, 3293], [3298, 3301], [3312, 3332],
|
||||
[3386, 3388], [3390, 3423], [3426, 3429], [3446, 3449], [3456, 3460], [3479, 3481],
|
||||
[3518, 3519], [3527, 3584], [3636, 3647], [3655, 3663], [3674, 3712], [3717, 3718],
|
||||
[3723, 3724], [3726, 3731], [3752, 3753], [3764, 3772], [3774, 3775], [3783, 3791],
|
||||
[3802, 3803], [3806, 3839], [3841, 3871], [3892, 3903], [3949, 3975], [3980, 4095],
|
||||
[4139, 4158], [4170, 4175], [4182, 4185], [4190, 4192], [4194, 4196], [4199, 4205],
|
||||
[4209, 4212], [4226, 4237], [4250, 4255], [4294, 4303], [4349, 4351], [4686, 4687],
|
||||
[4702, 4703], [4750, 4751], [4790, 4791], [4806, 4807], [4886, 4887], [4955, 4968],
|
||||
[4989, 4991], [5008, 5023], [5109, 5120], [5741, 5742], [5787, 5791], [5867, 5869],
|
||||
[5873, 5887], [5906, 5919], [5938, 5951], [5970, 5983], [6001, 6015], [6068, 6102],
|
||||
[6104, 6107], [6109, 6111], [6122, 6127], [6138, 6159], [6170, 6175], [6264, 6271],
|
||||
[6315, 6319], [6390, 6399], [6429, 6469], [6510, 6511], [6517, 6527], [6572, 6592],
|
||||
[6600, 6607], [6619, 6655], [6679, 6687], [6741, 6783], [6794, 6799], [6810, 6822],
|
||||
[6824, 6916], [6964, 6980], [6988, 6991], [7002, 7042], [7073, 7085], [7098, 7167],
|
||||
[7204, 7231], [7242, 7244], [7294, 7400], [7410, 7423], [7616, 7679], [7958, 7959],
|
||||
[7966, 7967], [8006, 8007], [8014, 8015], [8062, 8063], [8127, 8129], [8141, 8143],
|
||||
[8148, 8149], [8156, 8159], [8173, 8177], [8189, 8303], [8306, 8307], [8314, 8318],
|
||||
[8330, 8335], [8341, 8449], [8451, 8454], [8456, 8457], [8470, 8472], [8478, 8483],
|
||||
[8506, 8507], [8512, 8516], [8522, 8525], [8586, 9311], [9372, 9449], [9472, 10101],
|
||||
[10132, 11263], [11493, 11498], [11503, 11516], [11518, 11519], [11558, 11567],
|
||||
[11622, 11630], [11632, 11647], [11671, 11679], [11743, 11822], [11824, 12292],
|
||||
[12296, 12320], [12330, 12336], [12342, 12343], [12349, 12352], [12439, 12444],
|
||||
[12544, 12548], [12590, 12592], [12687, 12689], [12694, 12703], [12728, 12783],
|
||||
[12800, 12831], [12842, 12880], [12896, 12927], [12938, 12976], [12992, 13311],
|
||||
[19894, 19967], [40908, 40959], [42125, 42191], [42238, 42239], [42509, 42511],
|
||||
[42540, 42559], [42592, 42593], [42607, 42622], [42648, 42655], [42736, 42774],
|
||||
[42784, 42785], [42889, 42890], [42893, 43002], [43043, 43055], [43062, 43071],
|
||||
[43124, 43137], [43188, 43215], [43226, 43249], [43256, 43258], [43260, 43263],
|
||||
[43302, 43311], [43335, 43359], [43389, 43395], [43443, 43470], [43482, 43519],
|
||||
[43561, 43583], [43596, 43599], [43610, 43615], [43639, 43641], [43643, 43647],
|
||||
[43698, 43700], [43703, 43704], [43710, 43711], [43715, 43738], [43742, 43967],
|
||||
[44003, 44015], [44026, 44031], [55204, 55215], [55239, 55242], [55292, 55295],
|
||||
[57344, 63743], [64046, 64047], [64110, 64111], [64218, 64255], [64263, 64274],
|
||||
[64280, 64284], [64434, 64466], [64830, 64847], [64912, 64913], [64968, 65007],
|
||||
[65020, 65135], [65277, 65295], [65306, 65312], [65339, 65344], [65371, 65381],
|
||||
[65471, 65473], [65480, 65481], [65488, 65489], [65496, 65497]];
|
||||
for (i = 0; i < ranges.length; i++) {
|
||||
start = ranges[i][0];
|
||||
end = ranges[i][1];
|
||||
for (j = start; j <= end; j++) {
|
||||
result[j] = true;
|
||||
}
|
||||
}
|
||||
return result;
|
||||
})();
|
||||
|
||||
function splitQuery(query) {
|
||||
var result = [];
|
||||
var start = -1;
|
||||
for (var i = 0; i < query.length; i++) {
|
||||
if (splitChars[query.charCodeAt(i)]) {
|
||||
if (start !== -1) {
|
||||
result.push(query.slice(start, i));
|
||||
start = -1;
|
||||
}
|
||||
} else if (start === -1) {
|
||||
start = i;
|
||||
}
|
||||
}
|
||||
if (start !== -1) {
|
||||
result.push(query.slice(start));
|
||||
}
|
||||
return result;
|
||||
}
|
||||
|
||||
|
||||
|
||||
|
||||
/**
|
||||
* Search Module
|
||||
*/
|
||||
var Search = {
|
||||
|
||||
_index : null,
|
||||
_queued_query : null,
|
||||
_pulse_status : -1,
|
||||
|
||||
init : function() {
|
||||
var params = $.getQueryParameters();
|
||||
if (params.q) {
|
||||
var query = params.q[0];
|
||||
$('input[name="q"]')[0].value = query;
|
||||
this.performSearch(query);
|
||||
}
|
||||
},
|
||||
|
||||
loadIndex : function(url) {
|
||||
$.ajax({type: "GET", url: url, data: null,
|
||||
dataType: "script", cache: true,
|
||||
complete: function(jqxhr, textstatus) {
|
||||
if (textstatus != "success") {
|
||||
document.getElementById("searchindexloader").src = url;
|
||||
}
|
||||
}});
|
||||
},
|
||||
|
||||
setIndex : function(index) {
|
||||
var q;
|
||||
this._index = index;
|
||||
if ((q = this._queued_query) !== null) {
|
||||
this._queued_query = null;
|
||||
Search.query(q);
|
||||
}
|
||||
},
|
||||
|
||||
hasIndex : function() {
|
||||
return this._index !== null;
|
||||
},
|
||||
|
||||
deferQuery : function(query) {
|
||||
this._queued_query = query;
|
||||
},
|
||||
|
||||
stopPulse : function() {
|
||||
this._pulse_status = 0;
|
||||
},
|
||||
|
||||
startPulse : function() {
|
||||
if (this._pulse_status >= 0)
|
||||
return;
|
||||
function pulse() {
|
||||
var i;
|
||||
Search._pulse_status = (Search._pulse_status + 1) % 4;
|
||||
var dotString = '';
|
||||
for (i = 0; i < Search._pulse_status; i++)
|
||||
dotString += '.';
|
||||
Search.dots.text(dotString);
|
||||
if (Search._pulse_status > -1)
|
||||
window.setTimeout(pulse, 500);
|
||||
}
|
||||
pulse();
|
||||
},
|
||||
|
||||
/**
|
||||
* perform a search for something (or wait until index is loaded)
|
||||
*/
|
||||
performSearch : function(query) {
|
||||
// create the required interface elements
|
||||
this.out = $('#search-results');
|
||||
this.title = $('<h2>' + _('Searching') + '</h2>').appendTo(this.out);
|
||||
this.dots = $('<span></span>').appendTo(this.title);
|
||||
this.status = $('<p style="display: none"></p>').appendTo(this.out);
|
||||
this.output = $('<ul class="search"/>').appendTo(this.out);
|
||||
|
||||
$('#search-progress').text(_('Preparing search...'));
|
||||
this.startPulse();
|
||||
|
||||
// index already loaded, the browser was quick!
|
||||
if (this.hasIndex())
|
||||
this.query(query);
|
||||
else
|
||||
this.deferQuery(query);
|
||||
},
|
||||
|
||||
/**
|
||||
* execute search (requires search index to be loaded)
|
||||
*/
|
||||
query : function(query) {
|
||||
var i;
|
||||
var stopwords = ["a","and","are","as","at","be","but","by","for","if","in","into","is","it","near","no","not","of","on","or","such","that","the","their","then","there","these","they","this","to","was","will","with"];
|
||||
|
||||
// stem the searchterms and add them to the correct list
|
||||
var stemmer = new Stemmer();
|
||||
var searchterms = [];
|
||||
var excluded = [];
|
||||
var hlterms = [];
|
||||
var tmp = splitQuery(query);
|
||||
var objectterms = [];
|
||||
for (i = 0; i < tmp.length; i++) {
|
||||
if (tmp[i] !== "") {
|
||||
objectterms.push(tmp[i].toLowerCase());
|
||||
}
|
||||
|
||||
if ($u.indexOf(stopwords, tmp[i].toLowerCase()) != -1 || tmp[i].match(/^\d+$/) ||
|
||||
tmp[i] === "") {
|
||||
// skip this "word"
|
||||
continue;
|
||||
}
|
||||
// stem the word
|
||||
var word = stemmer.stemWord(tmp[i].toLowerCase());
|
||||
var toAppend;
|
||||
// select the correct list
|
||||
if (word[0] == '-') {
|
||||
toAppend = excluded;
|
||||
word = word.substr(1);
|
||||
}
|
||||
else {
|
||||
toAppend = searchterms;
|
||||
hlterms.push(tmp[i].toLowerCase());
|
||||
}
|
||||
// only add if not already in the list
|
||||
if (!$u.contains(toAppend, word))
|
||||
toAppend.push(word);
|
||||
}
|
||||
var highlightstring = '?highlight=' + $.urlencode(hlterms.join(" "));
|
||||
|
||||
// console.debug('SEARCH: searching for:');
|
||||
// console.info('required: ', searchterms);
|
||||
// console.info('excluded: ', excluded);
|
||||
|
||||
// prepare search
|
||||
var terms = this._index.terms;
|
||||
var titleterms = this._index.titleterms;
|
||||
|
||||
// array of [filename, title, anchor, descr, score]
|
||||
var results = [];
|
||||
$('#search-progress').empty();
|
||||
|
||||
// lookup as object
|
||||
for (i = 0; i < objectterms.length; i++) {
|
||||
var others = [].concat(objectterms.slice(0, i),
|
||||
objectterms.slice(i+1, objectterms.length));
|
||||
results = results.concat(this.performObjectSearch(objectterms[i], others));
|
||||
}
|
||||
|
||||
// lookup as search terms in fulltext
|
||||
results = results.concat(this.performTermsSearch(searchterms, excluded, terms, titleterms));
|
||||
|
||||
// let the scorer override scores with a custom scoring function
|
||||
if (Scorer.score) {
|
||||
for (i = 0; i < results.length; i++)
|
||||
results[i][4] = Scorer.score(results[i]);
|
||||
}
|
||||
|
||||
// now sort the results by score (in opposite order of appearance, since the
|
||||
// display function below uses pop() to retrieve items) and then
|
||||
// alphabetically
|
||||
results.sort(function(a, b) {
|
||||
var left = a[4];
|
||||
var right = b[4];
|
||||
if (left > right) {
|
||||
return 1;
|
||||
} else if (left < right) {
|
||||
return -1;
|
||||
} else {
|
||||
// same score: sort alphabetically
|
||||
left = a[1].toLowerCase();
|
||||
right = b[1].toLowerCase();
|
||||
return (left > right) ? -1 : ((left < right) ? 1 : 0);
|
||||
}
|
||||
});
|
||||
|
||||
// for debugging
|
||||
//Search.lastresults = results.slice(); // a copy
|
||||
//console.info('search results:', Search.lastresults);
|
||||
|
||||
// print the results
|
||||
var resultCount = results.length;
|
||||
function displayNextItem() {
|
||||
// results left, load the summary and display it
|
||||
if (results.length) {
|
||||
var item = results.pop();
|
||||
var listItem = $('<li style="display:none"></li>');
|
||||
if (DOCUMENTATION_OPTIONS.FILE_SUFFIX === '') {
|
||||
// dirhtml builder
|
||||
var dirname = item[0] + '/';
|
||||
if (dirname.match(/\/index\/$/)) {
|
||||
dirname = dirname.substring(0, dirname.length-6);
|
||||
} else if (dirname == 'index/') {
|
||||
dirname = '';
|
||||
}
|
||||
listItem.append($('<a/>').attr('href',
|
||||
DOCUMENTATION_OPTIONS.URL_ROOT + dirname +
|
||||
highlightstring + item[2]).html(item[1]));
|
||||
} else {
|
||||
// normal html builders
|
||||
listItem.append($('<a/>').attr('href',
|
||||
item[0] + DOCUMENTATION_OPTIONS.FILE_SUFFIX +
|
||||
highlightstring + item[2]).html(item[1]));
|
||||
}
|
||||
if (item[3]) {
|
||||
listItem.append($('<span> (' + item[3] + ')</span>'));
|
||||
Search.output.append(listItem);
|
||||
listItem.slideDown(5, function() {
|
||||
displayNextItem();
|
||||
});
|
||||
} else if (DOCUMENTATION_OPTIONS.HAS_SOURCE) {
|
||||
$.ajax({url: DOCUMENTATION_OPTIONS.URL_ROOT + '_sources/' + item[0] + '.md.txt',
|
||||
dataType: "text",
|
||||
complete: function(jqxhr, textstatus) {
|
||||
var data = jqxhr.responseText;
|
||||
if (data !== '' && data !== undefined) {
|
||||
listItem.append(Search.makeSearchSummary(data, searchterms, hlterms));
|
||||
}
|
||||
Search.output.append(listItem);
|
||||
listItem.slideDown(5, function() {
|
||||
displayNextItem();
|
||||
});
|
||||
}});
|
||||
} else {
|
||||
// no source available, just display title
|
||||
Search.output.append(listItem);
|
||||
listItem.slideDown(5, function() {
|
||||
displayNextItem();
|
||||
});
|
||||
}
|
||||
}
|
||||
// search finished, update title and status message
|
||||
else {
|
||||
Search.stopPulse();
|
||||
Search.title.text(_('Search Results'));
|
||||
if (!resultCount)
|
||||
Search.status.text(_('Your search did not match any documents. Please make sure that all words are spelled correctly and that you\'ve selected enough categories.'));
|
||||
else
|
||||
Search.status.text(_('Search finished, found %s page(s) matching the search query.').replace('%s', resultCount));
|
||||
Search.status.fadeIn(500);
|
||||
}
|
||||
}
|
||||
displayNextItem();
|
||||
},
|
||||
|
||||
/**
|
||||
* search for object names
|
||||
*/
|
||||
performObjectSearch : function(object, otherterms) {
|
||||
var filenames = this._index.docnames;
|
||||
var objects = this._index.objects;
|
||||
var objnames = this._index.objnames;
|
||||
var titles = this._index.titles;
|
||||
|
||||
var i;
|
||||
var results = [];
|
||||
|
||||
for (var prefix in objects) {
|
||||
for (var name in objects[prefix]) {
|
||||
var fullname = (prefix ? prefix + '.' : '') + name;
|
||||
if (fullname.toLowerCase().indexOf(object) > -1) {
|
||||
var score = 0;
|
||||
var parts = fullname.split('.');
|
||||
// check for different match types: exact matches of full name or
|
||||
// "last name" (i.e. last dotted part)
|
||||
if (fullname == object || parts[parts.length - 1] == object) {
|
||||
score += Scorer.objNameMatch;
|
||||
// matches in last name
|
||||
} else if (parts[parts.length - 1].indexOf(object) > -1) {
|
||||
score += Scorer.objPartialMatch;
|
||||
}
|
||||
var match = objects[prefix][name];
|
||||
var objname = objnames[match[1]][2];
|
||||
var title = titles[match[0]];
|
||||
// If more than one term searched for, we require other words to be
|
||||
// found in the name/title/description
|
||||
if (otherterms.length > 0) {
|
||||
var haystack = (prefix + ' ' + name + ' ' +
|
||||
objname + ' ' + title).toLowerCase();
|
||||
var allfound = true;
|
||||
for (i = 0; i < otherterms.length; i++) {
|
||||
if (haystack.indexOf(otherterms[i]) == -1) {
|
||||
allfound = false;
|
||||
break;
|
||||
}
|
||||
}
|
||||
if (!allfound) {
|
||||
continue;
|
||||
}
|
||||
}
|
||||
var descr = objname + _(', in ') + title;
|
||||
|
||||
var anchor = match[3];
|
||||
if (anchor === '')
|
||||
anchor = fullname;
|
||||
else if (anchor == '-')
|
||||
anchor = objnames[match[1]][1] + '-' + fullname;
|
||||
// add custom score for some objects according to scorer
|
||||
if (Scorer.objPrio.hasOwnProperty(match[2])) {
|
||||
score += Scorer.objPrio[match[2]];
|
||||
} else {
|
||||
score += Scorer.objPrioDefault;
|
||||
}
|
||||
results.push([filenames[match[0]], fullname, '#'+anchor, descr, score]);
|
||||
}
|
||||
}
|
||||
}
|
||||
|
||||
return results;
|
||||
},
|
||||
|
||||
/**
|
||||
* search for full-text terms in the index
|
||||
*/
|
||||
performTermsSearch : function(searchterms, excluded, terms, titleterms) {
|
||||
var filenames = this._index.docnames;
|
||||
var titles = this._index.titles;
|
||||
|
||||
var i, j, file;
|
||||
var fileMap = {};
|
||||
var scoreMap = {};
|
||||
var results = [];
|
||||
|
||||
// perform the search on the required terms
|
||||
for (i = 0; i < searchterms.length; i++) {
|
||||
var word = searchterms[i];
|
||||
var files = [];
|
||||
var _o = [
|
||||
{files: terms[word], score: Scorer.term},
|
||||
{files: titleterms[word], score: Scorer.title}
|
||||
];
|
||||
|
||||
// no match but word was a required one
|
||||
if ($u.every(_o, function(o){return o.files === undefined;})) {
|
||||
break;
|
||||
}
|
||||
// found search word in contents
|
||||
$u.each(_o, function(o) {
|
||||
var _files = o.files;
|
||||
if (_files === undefined)
|
||||
return
|
||||
|
||||
if (_files.length === undefined)
|
||||
_files = [_files];
|
||||
files = files.concat(_files);
|
||||
|
||||
// set score for the word in each file to Scorer.term
|
||||
for (j = 0; j < _files.length; j++) {
|
||||
file = _files[j];
|
||||
if (!(file in scoreMap))
|
||||
scoreMap[file] = {}
|
||||
scoreMap[file][word] = o.score;
|
||||
}
|
||||
});
|
||||
|
||||
// create the mapping
|
||||
for (j = 0; j < files.length; j++) {
|
||||
file = files[j];
|
||||
if (file in fileMap)
|
||||
fileMap[file].push(word);
|
||||
else
|
||||
fileMap[file] = [word];
|
||||
}
|
||||
}
|
||||
|
||||
// now check if the files don't contain excluded terms
|
||||
for (file in fileMap) {
|
||||
var valid = true;
|
||||
|
||||
// check if all requirements are matched
|
||||
if (fileMap[file].length != searchterms.length)
|
||||
continue;
|
||||
|
||||
// ensure that none of the excluded terms is in the search result
|
||||
for (i = 0; i < excluded.length; i++) {
|
||||
if (terms[excluded[i]] == file ||
|
||||
titleterms[excluded[i]] == file ||
|
||||
$u.contains(terms[excluded[i]] || [], file) ||
|
||||
$u.contains(titleterms[excluded[i]] || [], file)) {
|
||||
valid = false;
|
||||
break;
|
||||
}
|
||||
}
|
||||
|
||||
// if we have still a valid result we can add it to the result list
|
||||
if (valid) {
|
||||
// select one (max) score for the file.
|
||||
// for better ranking, we should calculate ranking by using words statistics like basic tf-idf...
|
||||
var score = $u.max($u.map(fileMap[file], function(w){return scoreMap[file][w]}));
|
||||
results.push([filenames[file], titles[file], '', null, score]);
|
||||
}
|
||||
}
|
||||
return results;
|
||||
},
|
||||
|
||||
/**
|
||||
* helper function to return a node containing the
|
||||
* search summary for a given text. keywords is a list
|
||||
* of stemmed words, hlwords is the list of normal, unstemmed
|
||||
* words. the first one is used to find the occurrence, the
|
||||
* latter for highlighting it.
|
||||
*/
|
||||
makeSearchSummary : function(text, keywords, hlwords) {
|
||||
var textLower = text.toLowerCase();
|
||||
var start = 0;
|
||||
$.each(keywords, function() {
|
||||
var i = textLower.indexOf(this.toLowerCase());
|
||||
if (i > -1)
|
||||
start = i;
|
||||
});
|
||||
start = Math.max(start - 120, 0);
|
||||
var excerpt = ((start > 0) ? '...' : '') +
|
||||
$.trim(text.substr(start, 240)) +
|
||||
((start + 240 - text.length) ? '...' : '');
|
||||
var rv = $('<div class="context"></div>').text(excerpt);
|
||||
$.each(hlwords, function() {
|
||||
rv = rv.highlightText(this, 'highlighted');
|
||||
});
|
||||
return rv;
|
||||
}
|
||||
};
|
||||
|
||||
/* Search initialization removed for Read the Docs */
|
||||
$(document).ready(function() {
|
||||
Search.init();
|
||||
});
|
||||
5
doc/_static/xgboost-theme/footer.html
vendored
5
doc/_static/xgboost-theme/footer.html
vendored
@ -1,5 +0,0 @@
|
||||
<div class="container">
|
||||
<div class="footer">
|
||||
<p> © 2015-2016 DMLC. All rights reserved. </p>
|
||||
</div>
|
||||
</div>
|
||||
58
doc/_static/xgboost-theme/index.html
vendored
58
doc/_static/xgboost-theme/index.html
vendored
@ -1,58 +0,0 @@
|
||||
<div class="splash">
|
||||
<div class="container">
|
||||
<div class="row">
|
||||
<div class="col-lg-12">
|
||||
<h1>Scalable and Flexible Gradient Boosting</h1>
|
||||
<div id="social">
|
||||
<span>
|
||||
<iframe src="https://ghbtns.com/github-btn.html?user=dmlc&repo=xgboost&type=star&count=true&v=2"
|
||||
frameborder="0" scrolling="0" width="120px" height="20px"></iframe>
|
||||
<iframe src="https://ghbtns.com/github-btn.html?user=dmlc&repo=xgboost&type=fork&count=true&v=2"
|
||||
frameborder="0" scrolling="0" width="100px" height="20px"></iframe>
|
||||
</span>
|
||||
</div> <!-- end of social -->
|
||||
<div class="get_start">
|
||||
<a href="get_started/" class="get_start_btn">Get Started</a>
|
||||
</div> <!-- end of get started button -->
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
|
||||
<div class="section-tout">
|
||||
<div class="container">
|
||||
<div class="row">
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-flag"></i> Flexible</h3>
|
||||
<p>Supports regression, classification, ranking and user defined objectives.
|
||||
</p>
|
||||
</div>
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-cube"></i> Portable</h3>
|
||||
<p>Runs on Windows, Linux and OS X, as well as various cloud Platforms</p>
|
||||
</div>
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-wrench"></i>Multiple Languages</h3>
|
||||
<p>Supports multiple languages including C++, Python, R, Java, Scala, Julia.</p>
|
||||
</div>
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-cogs"></i> Battle-tested</h3>
|
||||
<p>Wins many data science and machine learning challenges.
|
||||
Used in production by multiple companies.
|
||||
</p>
|
||||
</div>
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-cloud"></i>Distributed on Cloud</h3>
|
||||
<p>Supports distributed training on multiple machines, including AWS,
|
||||
GCE, Azure, and Yarn clusters. Can be integrated with Flink, Spark and other cloud dataflow systems.</p>
|
||||
</div>
|
||||
<div class="col-lg-4 col-sm-6">
|
||||
<h3><i class="fa fa-rocket"></i> Performance</h3>
|
||||
<p>The well-optimized backend system for the best performance with limited resources.
|
||||
The distributed version solves problems beyond billions of examples with same code.
|
||||
</p>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
156
doc/_static/xgboost-theme/layout.html
vendored
156
doc/_static/xgboost-theme/layout.html
vendored
@ -1,156 +0,0 @@
|
||||
{%- block doctype -%}
|
||||
<!DOCTYPE html>
|
||||
{%- endblock %}
|
||||
{%- set reldelim1 = reldelim1 is not defined and ' »' or reldelim1 %}
|
||||
{%- set reldelim2 = reldelim2 is not defined and ' |' or reldelim2 %}
|
||||
{%- set render_sidebar = (not embedded) and (not theme_nosidebar|tobool) and
|
||||
(sidebars != []) %}
|
||||
{%- set url_root = pathto('', 1) %}
|
||||
{%- if url_root == '#' %}{% set url_root = '' %}{% endif %}
|
||||
{%- if not embedded and docstitle %}
|
||||
{%- set titlesuffix = " — "|safe + docstitle|e %}
|
||||
{%- else %}
|
||||
{%- set titlesuffix = "" %}
|
||||
{%- endif %}
|
||||
|
||||
{%- macro searchform(classes, button) %}
|
||||
<form class="{{classes}}" role="search" action="{{ pathto('search') }}" method="get">
|
||||
<div class="form-group">
|
||||
<input type="text" name="q" class="form-control" {{ 'placeholder="Search"' if not button }} >
|
||||
</div>
|
||||
<input type="hidden" name="check_keywords" value="yes" />
|
||||
<input type="hidden" name="area" value="default" />
|
||||
{% if button %}
|
||||
<input type="submit" class="btn btn-default" value="search">
|
||||
{% endif %}
|
||||
</form>
|
||||
{%- endmacro %}
|
||||
|
||||
{%- macro sidebarglobal() %}
|
||||
<ul class="globaltoc">
|
||||
{{ toctree(maxdepth=2|toint, collapse=False,includehidden=theme_globaltoc_includehidden|tobool) }}
|
||||
</ul>
|
||||
{%- endmacro %}
|
||||
|
||||
{%- macro sidebar() %}
|
||||
{%- if render_sidebar %}
|
||||
<div class="sphinxsidebar" role="navigation" aria-label="main navigation">
|
||||
<div class="sphinxsidebarwrapper">
|
||||
{%- block sidebartoc %}
|
||||
{%- include "localtoc.html" %}
|
||||
{%- endblock %}
|
||||
</div>
|
||||
</div>
|
||||
{%- endif %}
|
||||
{%- endmacro %}
|
||||
|
||||
|
||||
{%- macro script() %}
|
||||
<script type="text/javascript">
|
||||
var DOCUMENTATION_OPTIONS = {
|
||||
URL_ROOT: '{{ url_root }}',
|
||||
VERSION: '{{ release|e }}',
|
||||
COLLAPSE_INDEX: false,
|
||||
FILE_SUFFIX: '{{ '' if no_search_suffix else file_suffix }}',
|
||||
HAS_SOURCE: {{ has_source|lower }}
|
||||
};
|
||||
</script>
|
||||
|
||||
{% for name in ['jquery.js', 'underscore.js', 'doctools.js', 'searchtools-new.js'] %}
|
||||
<script type="text/javascript" src="{{ pathto('_static/' + name, 1) }}"></script>
|
||||
{% endfor %}
|
||||
|
||||
<script type="text/javascript" src="https://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"></script>
|
||||
|
||||
<!-- {%- for scriptfile in script_files %} -->
|
||||
<!-- <script type="text/javascript" src="{{ pathto(scriptfile, 1) }}"></script> -->
|
||||
<!-- {%- endfor %} -->
|
||||
{%- endmacro %}
|
||||
|
||||
{%- macro css() %}
|
||||
<link rel="stylesheet" href="https://maxcdn.bootstrapcdn.com/bootstrap/3.3.6/css/bootstrap.min.css" integrity="sha384-1q8mTJOASx8j1Au+a5WDVnPi2lkFfwwEAa8hDDdjZlpLegxhjVME1fgjWPGmkzs7" crossorigin="anonymous">
|
||||
{% if pagename == 'index' %}
|
||||
<link rel="stylesheet" href="https://maxcdn.bootstrapcdn.com/font-awesome/4.5.0/css/font-awesome.min.css">
|
||||
{%- else %}
|
||||
<link rel="stylesheet" href="{{ pathto('_static/' + style, 1) }}" type="text/css" />
|
||||
<link rel="stylesheet" href="{{ pathto('_static/pygments.css', 1) }}" type="text/css" />
|
||||
{%- endif %}
|
||||
|
||||
<link rel="stylesheet" href="{{ pathto('_static/xgboost.css', 1) }}">
|
||||
{%- endmacro %}
|
||||
|
||||
<html lang="en">
|
||||
<head>
|
||||
<meta charset="{{ encoding }}">
|
||||
<meta http-equiv="X-UA-Compatible" content="IE=edge">
|
||||
<meta name="viewport" content="width=device-width, initial-scale=1">
|
||||
{# The above 3 meta tags *must* come first in the head; any other head content
|
||||
must come *after* these tags. #}
|
||||
{{ metatags }}
|
||||
{%- block htmltitle %}
|
||||
{%- if pagename != 'index' %}
|
||||
<title>{{ title|striptags|e }}{{ titlesuffix }}</title>
|
||||
{%- else %}
|
||||
<title>XGBoost Documents</title>
|
||||
{%- endif %}
|
||||
{%- endblock %}
|
||||
{{ css() }}
|
||||
{%- if not embedded %}
|
||||
{{ script() }}
|
||||
{%- if use_opensearch %}
|
||||
<link rel="search" type="application/opensearchdescription+xml"
|
||||
title="{% trans docstitle=docstitle|e %}Search within {{ docstitle }}{% endtrans %}"
|
||||
href="{{ pathto('_static/opensearch.xml', 1) }}"/>
|
||||
{%- endif %}
|
||||
{%- if favicon %}
|
||||
<link rel="shortcut icon" href="{{ pathto('_static/' + favicon, 1) }}"/>
|
||||
{%- endif %}
|
||||
{%- endif %}
|
||||
{%- block linktags %}
|
||||
{%- if hasdoc('about') %}
|
||||
<link rel="author" title="{{ _('About these documents') }}" href="{{ pathto('about') }}" />
|
||||
{%- endif %}
|
||||
{%- if hasdoc('genindex') %}
|
||||
<link rel="index" title="{{ _('Index') }}" href="{{ pathto('genindex') }}" />
|
||||
{%- endif %}
|
||||
{%- if hasdoc('search') %}
|
||||
<link rel="search" title="{{ _('Search') }}" href="{{ pathto('search') }}" />
|
||||
{%- endif %}
|
||||
{%- if hasdoc('copyright') %}
|
||||
<link rel="copyright" title="{{ _('Copyright') }}" href="{{ pathto('copyright') }}" />
|
||||
{%- endif %}
|
||||
{%- if parents %}
|
||||
<link rel="up" title="{{ parents[-1].title|striptags|e }}" href="{{ parents[-1].link|e }}" />
|
||||
{%- endif %}
|
||||
{%- if next %}
|
||||
<link rel="next" title="{{ next.title|striptags|e }}" href="{{ next.link|e }}" />
|
||||
{%- endif %}
|
||||
{%- if prev %}
|
||||
<link rel="prev" title="{{ prev.title|striptags|e }}" href="{{ prev.link|e }}" />
|
||||
{%- endif %}
|
||||
{%- endblock %}
|
||||
{%- block extrahead %} {% endblock %}
|
||||
|
||||
<link rel="icon" type="image/png" href="https://raw.githubusercontent.com/dmlc/web-data/master/mxnet/image/mxnet-icon.png">
|
||||
</head>
|
||||
<body role="document">
|
||||
{%- include "navbar.html" %}
|
||||
|
||||
{% if pagename != 'index' %}
|
||||
<div class="container">
|
||||
<div class="row">
|
||||
{{ sidebar() }}
|
||||
<div class="content">
|
||||
{% block body %} {% endblock %}
|
||||
{%- include "footer.html" %}
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
{%- else %}
|
||||
{%- include "index.html" %}
|
||||
{%- include "footer.html" %}
|
||||
{%- endif %} <!-- pagename != index -->
|
||||
|
||||
<script src="https://maxcdn.bootstrapcdn.com/bootstrap/3.3.6/js/bootstrap.min.js" integrity="sha384-0mSbJDEHialfmuBBQP6A4Qrprq5OVfW37PRR3j5ELqxss1yVqOtnepnHVP9aJ7xS" crossorigin="anonymous"></script>
|
||||
</body>
|
||||
</html>
|
||||
41
doc/_static/xgboost-theme/navbar.html
vendored
41
doc/_static/xgboost-theme/navbar.html
vendored
@ -1,41 +0,0 @@
|
||||
<div class="navbar navbar-default navbar-fixed-top">
|
||||
<div class="container">
|
||||
<div class="navbar-header">
|
||||
<button type="button" class="navbar-toggle collapsed" data-toggle="collapse" data-target="#navbar" aria-expanded="false" aria-controls="navbar">
|
||||
<span class="sr-only">Toggle navigation</span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
<span class="icon-bar"></span>
|
||||
</button>
|
||||
</div>
|
||||
<div id="navbar" class="navbar-collapse collapse">
|
||||
<ul id="navbar" class="navbar navbar-left">
|
||||
<li> <a href="{{url_root}}">XGBoost</a> </li>
|
||||
{% for name in ['Get Started', 'Tutorials', 'How To'] %}
|
||||
<li> <a href="{{url_root}}{{name.lower()|replace(" ", "_")}}/index.html">{{name}}</a> </li>
|
||||
{% endfor %}
|
||||
{% for name in ['Packages'] %}
|
||||
<li class="dropdown">
|
||||
<a href="#" class="dropdown-toggle" data-toggle="dropdown" role="button" aria-haspopup="true" aria-expanded="true">{{name}} <span class="caret"></span></a>
|
||||
<ul class="dropdown-menu">
|
||||
<li><a href="{{url_root}}python/index.html">Python</a></li>
|
||||
<li><a href="{{url_root}}R-package/index.html">R</a></li>
|
||||
<li><a href="{{url_root}}jvm/index.html">JVM</a></li>
|
||||
<li><a href="{{url_root}}julia/index.html">Julia</a></li>
|
||||
<li><a href="{{url_root}}cli/index.html">CLI</a></li>
|
||||
<li><a href="{{url_root}}gpu/index.html">GPU</a></li>
|
||||
</ul>
|
||||
</li>
|
||||
{% endfor %}
|
||||
<li> <a href="{{url_root}}/parameter.html"> Knobs </a> </li>
|
||||
<li> {{searchform('', False)}} </li>
|
||||
</ul>
|
||||
<!--
|
||||
<ul id="navbar" class="navbar navbar-right">
|
||||
<li> <a href="{{url_root}}index.html"><span class="flag-icon flag-icon-us"></span></a> </li>
|
||||
<li> <a href="{{url_root}}/zh/index.html"><span class="flag-icon flag-icon-cn"></span></a> </li>
|
||||
</ul>
|
||||
navbar -->
|
||||
</div>
|
||||
</div>
|
||||
</div>
|
||||
2
doc/_static/xgboost-theme/theme.conf
vendored
2
doc/_static/xgboost-theme/theme.conf
vendored
@ -1,2 +0,0 @@
|
||||
[theme]
|
||||
inherit = basic
|
||||
232
doc/_static/xgboost.css
vendored
232
doc/_static/xgboost.css
vendored
@ -1,232 +0,0 @@
|
||||
/* header section */
|
||||
.splash{
|
||||
padding:5em 0 1em 0;
|
||||
background-color:#0079b2;
|
||||
/* background-image:url(../img/bg.jpg); */
|
||||
background-size:cover;
|
||||
background-attachment:fixed;
|
||||
color:#fff;
|
||||
text-align:center
|
||||
}
|
||||
|
||||
.splash h1{
|
||||
font-size: 40px;
|
||||
margin-bottom: 20px;
|
||||
}
|
||||
.splash .social{
|
||||
margin:2em 0
|
||||
}
|
||||
|
||||
.splash .get_start {
|
||||
margin:2em 0
|
||||
}
|
||||
|
||||
.splash .get_start_btn {
|
||||
border: 2px solid #FFFFFF;
|
||||
border-radius: 5px;
|
||||
color: #FFFFFF;
|
||||
display: inline-block;
|
||||
font-size: 26px;
|
||||
padding: 9px 20px;
|
||||
}
|
||||
|
||||
.section-tout{
|
||||
padding:3em 0 3em;
|
||||
border-bottom:1px solid rgba(0,0,0,.05);
|
||||
background-color:#eaf1f1
|
||||
}
|
||||
.section-tout .fa{
|
||||
margin-right:.5em
|
||||
}
|
||||
|
||||
.section-tout h3{
|
||||
font-size:20px;
|
||||
}
|
||||
|
||||
.section-tout p {
|
||||
margin-bottom:2em
|
||||
}
|
||||
|
||||
.section-inst{
|
||||
padding:3em 0 3em;
|
||||
border-bottom:1px solid rgba(0,0,0,.05);
|
||||
|
||||
text-align:center
|
||||
}
|
||||
|
||||
.section-inst p {
|
||||
margin-bottom:2em
|
||||
}
|
||||
.section-inst img {
|
||||
-webkit-filter: grayscale(90%); /* Chrome, Safari, Opera */
|
||||
filter: grayscale(90%);
|
||||
margin-bottom:2em
|
||||
}
|
||||
.section-inst img:hover {
|
||||
-webkit-filter: grayscale(0%); /* Chrome, Safari, Opera */
|
||||
filter: grayscale(0%);
|
||||
}
|
||||
|
||||
.footer{
|
||||
padding-top: 40px;
|
||||
}
|
||||
.footer li{
|
||||
float:right;
|
||||
margin-right:1.5em;
|
||||
margin-bottom:1.5em
|
||||
}
|
||||
.footer p{
|
||||
font-size: 15px;
|
||||
color: #888;
|
||||
clear:right;
|
||||
margin-bottom:0
|
||||
}
|
||||
|
||||
|
||||
/* sidebar */
|
||||
div.sphinxsidebar {
|
||||
margin-top: 20px;
|
||||
margin-left: 0;
|
||||
position: fixed;
|
||||
overflow-y: scroll;
|
||||
width: 250px;
|
||||
top: 52px;
|
||||
bottom: 0;
|
||||
display: none
|
||||
}
|
||||
div.sphinxsidebar ul { padding: 0 }
|
||||
div.sphinxsidebar ul ul { margin-left: 15px }
|
||||
|
||||
@media (min-width:1200px) {
|
||||
.content { float: right; width: 66.66666667%; margin-right: 5% }
|
||||
div.sphinxsidebar {display: block}
|
||||
}
|
||||
|
||||
|
||||
.github-btn { border: 0; overflow: hidden }
|
||||
|
||||
.container {
|
||||
margin-right: auto;
|
||||
margin-left: auto;
|
||||
padding-left: 15px;
|
||||
padding-right: 15px
|
||||
}
|
||||
|
||||
body>.container {
|
||||
padding-top: 80px
|
||||
}
|
||||
|
||||
body {
|
||||
font-size: 16px;
|
||||
}
|
||||
|
||||
pre {
|
||||
font-size: 14px;
|
||||
}
|
||||
|
||||
/* navbar */
|
||||
.navbar {
|
||||
background-color:#0079b2;
|
||||
border: 0px;
|
||||
height: 65px;
|
||||
}
|
||||
.navbar-right li {
|
||||
display:inline-block;
|
||||
vertical-align:top;
|
||||
padding: 22px 4px;
|
||||
}
|
||||
|
||||
.navbar-left li {
|
||||
display:inline-block;
|
||||
vertical-align:top;
|
||||
padding: 17px 10px;
|
||||
/* margin: 0 5px; */
|
||||
}
|
||||
|
||||
.navbar-left li a {
|
||||
font-size: 22px;
|
||||
color: #fff;
|
||||
}
|
||||
|
||||
.navbar-left > li > a:hover{
|
||||
color:#fff;
|
||||
}
|
||||
.flag-icon {
|
||||
background-size: contain;
|
||||
background-position: 50%;
|
||||
background-repeat: no-repeat;
|
||||
position: relative;
|
||||
display: inline-block;
|
||||
width: 1.33333333em;
|
||||
line-height: 1em;
|
||||
}
|
||||
|
||||
.flag-icon:before {
|
||||
content: "\00a0";
|
||||
}
|
||||
|
||||
|
||||
.flag-icon-cn {
|
||||
background-image: url(./cn.svg);
|
||||
}
|
||||
|
||||
.flag-icon-us {
|
||||
background-image: url(./us.svg);
|
||||
}
|
||||
|
||||
|
||||
/* .flags { */
|
||||
/* padding: 10px; */
|
||||
/* } */
|
||||
|
||||
.navbar-brand >img {
|
||||
width: 110px;
|
||||
}
|
||||
|
||||
.dropdown-menu li {
|
||||
padding: 0px 0px;
|
||||
width: 100%;
|
||||
}
|
||||
.dropdown-menu li a {
|
||||
color: #0079b2;
|
||||
font-size: 20px;
|
||||
}
|
||||
|
||||
.section h1 {
|
||||
padding-top: 90px;
|
||||
margin-top: -60px;
|
||||
padding-bottom: 10px;
|
||||
font-size: 28px;
|
||||
}
|
||||
|
||||
.section h2 {
|
||||
padding-top: 80px;
|
||||
margin-top: -60px;
|
||||
padding-bottom: 10px;
|
||||
font-size: 22px;
|
||||
}
|
||||
|
||||
.section h3 {
|
||||
padding-top: 80px;
|
||||
margin-top: -64px;
|
||||
padding-bottom: 8px;
|
||||
}
|
||||
|
||||
.section h4 {
|
||||
padding-top: 80px;
|
||||
margin-top: -64px;
|
||||
padding-bottom: 8px;
|
||||
}
|
||||
|
||||
dt {
|
||||
margin-top: -76px;
|
||||
padding-top: 76px;
|
||||
}
|
||||
|
||||
dt:target, .highlighted {
|
||||
background-color: #fff;
|
||||
}
|
||||
|
||||
.section code.descname {
|
||||
font-size: 1em;
|
||||
}
|
||||
399
doc/build.md
399
doc/build.md
@ -1,399 +0,0 @@
|
||||
Installation Guide
|
||||
==================
|
||||
|
||||
**NOTE**. If you are planning to use Python on a Linux system, consider installing XGBoost from a pre-built binary wheel. The wheel is available from Python Package Index (PyPI). You may download and install it by running
|
||||
```bash
|
||||
# Ensure that you are downloading xgboost-{version}-py2.py3-none-manylinux1_x86_64.whl
|
||||
pip3 install xgboost
|
||||
```
|
||||
* This package will support GPU algorithms (`gpu_exact`, `gpu_hist`) on machines with NVIDIA GPUs.
|
||||
* Currently, PyPI has a binary wheel only for 64-bit Linux.
|
||||
|
||||
# Building XGBoost from source
|
||||
This page gives instructions on how to build and install the xgboost package from
|
||||
scratch on various systems. It consists of two steps:
|
||||
|
||||
1. First build the shared library from the C++ codes (`libxgboost.so` for Linux/OSX and `xgboost.dll` for Windows).
|
||||
- Exception: for R-package installation please directly refer to the R package section.
|
||||
2. Then install the language packages (e.g. Python Package).
|
||||
|
||||
***Important*** the newest version of xgboost uses submodule to maintain packages. So when you clone the repo, remember to use the recursive option as follows.
|
||||
```bash
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
```
|
||||
For windows users who use github tools, you can open the git shell, and type the following command.
|
||||
```bash
|
||||
git submodule init
|
||||
git submodule update
|
||||
```
|
||||
|
||||
Please refer to [Trouble Shooting Section](#trouble-shooting) first if you had any problem
|
||||
during installation. If the instructions do not work for you, please feel free
|
||||
to ask questions at [xgboost/issues](https://github.com/dmlc/xgboost/issues), or
|
||||
even better to send pull request if you can fix the problem.
|
||||
|
||||
## Contents
|
||||
- [Build the Shared Library](#build-the-shared-library)
|
||||
- [Building on Ubuntu/Debian](#building-on-ubuntu-debian)
|
||||
- [Building on macOS](#building-on-macos)
|
||||
- [Building on Windows](#building-on-windows)
|
||||
- [Building with GPU support](#building-with-gpu-support)
|
||||
- [Windows Binaries](#windows-binaries)
|
||||
- [Customized Building](#customized-building)
|
||||
- [Python Package Installation](#python-package-installation)
|
||||
- [R Package Installation](#r-package-installation)
|
||||
- [Trouble Shooting](#trouble-shooting)
|
||||
|
||||
## Build the Shared Library
|
||||
|
||||
Our goal is to build the shared library:
|
||||
- On Linux/OSX the target library is `libxgboost.so`
|
||||
- On Windows the target library is `xgboost.dll`
|
||||
|
||||
The minimal building requirement is
|
||||
|
||||
- A recent c++ compiler supporting C++ 11 (g++-4.8 or higher)
|
||||
|
||||
We can edit `make/config.mk` to change the compile options, and then build by
|
||||
`make`. If everything goes well, we can go to the specific language installation section.
|
||||
|
||||
### Building on Ubuntu/Debian
|
||||
|
||||
On Ubuntu, one builds xgboost by
|
||||
|
||||
```bash
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost; make -j4
|
||||
```
|
||||
|
||||
### Building on macOS
|
||||
|
||||
**Install with pip - simple method**
|
||||
|
||||
First, make sure you obtained *gcc-5* (newer version does not work with this method yet). Note: installation of `gcc` can take a while (~ 30 minutes)
|
||||
|
||||
```bash
|
||||
brew install gcc5
|
||||
```
|
||||
|
||||
You might need to run the following command with `sudo` if you run into some permission errors:
|
||||
|
||||
```bash
|
||||
pip install xgboost
|
||||
```
|
||||
|
||||
**Build from the source code - advanced method**
|
||||
|
||||
First, obtain gcc-7.x.x with brew (https://brew.sh/) if you want multi-threaded version, otherwise, Clang is ok if OpenMP / multi-threaded is not required. Note: installation of `gcc` can take a while (~ 30 minutes)
|
||||
|
||||
```bash
|
||||
brew install gcc
|
||||
```
|
||||
|
||||
Now, clone the repository
|
||||
|
||||
```bash
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost; cp make/config.mk ./config.mk
|
||||
```
|
||||
|
||||
Open config.mk and uncomment these two lines
|
||||
|
||||
```config.mk
|
||||
export CC = gcc
|
||||
export CXX = g++
|
||||
```
|
||||
|
||||
and replace these two lines into(5 or 6 or 7; depending on your gcc-version)
|
||||
|
||||
```config.mk
|
||||
export CC = gcc-7
|
||||
export CXX = g++-7
|
||||
```
|
||||
|
||||
To find your gcc version
|
||||
|
||||
```bash
|
||||
gcc-version
|
||||
```
|
||||
|
||||
and build using the following commands
|
||||
|
||||
```bash
|
||||
make -j4
|
||||
```
|
||||
head over to `Python Package Installation` for the next steps
|
||||
|
||||
### Building on Windows
|
||||
You need to first clone the xgboost repo with recursive option clone the submodules.
|
||||
If you are using github tools, you can open the git-shell, and type the following command.
|
||||
We recommend using [Git for Windows](https://git-for-windows.github.io/)
|
||||
because it brings a standard bash shell. This will highly ease the installation process.
|
||||
|
||||
```bash
|
||||
git submodule init
|
||||
git submodule update
|
||||
```
|
||||
|
||||
XGBoost support both build by MSVC or MinGW. Here is how you can build xgboost library using MinGW.
|
||||
|
||||
After installing [Git for Windows](https://git-for-windows.github.io/), you should have a shortcut `Git Bash`.
|
||||
All the following steps are in the `Git Bash`.
|
||||
|
||||
In MinGW, `make` command comes with the name `mingw32-make`. You can add the following line into the `.bashrc` file.
|
||||
```bash
|
||||
alias make='mingw32-make'
|
||||
```
|
||||
(On 64-bit Windows, you should get [mingw64](https://sourceforge.net/projects/mingw-w64/) instead.) Make sure
|
||||
that the path to MinGW is in the system PATH.
|
||||
|
||||
To build with MinGW, type:
|
||||
|
||||
```bash
|
||||
cp make/mingw64.mk config.mk; make -j4
|
||||
```
|
||||
|
||||
To build with Visual Studio 2013 use cmake. Make sure you have a recent version of cmake added to your path and then from the xgboost directory:
|
||||
|
||||
```bash
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -G"Visual Studio 12 2013 Win64"
|
||||
```
|
||||
|
||||
This specifies an out of source build using the MSVC 12 64 bit generator. Open the .sln file in the build directory and build with Visual Studio. To use the Python module you can copy `xgboost.dll` into python-package\xgboost.
|
||||
|
||||
Other versions of Visual Studio may work but are untested.
|
||||
|
||||
### Building with GPU support
|
||||
|
||||
Linux users may simply install prebuilt python binaries:
|
||||
```bash
|
||||
pip install xgboost
|
||||
```
|
||||
|
||||
XGBoost can be built from source with GPU support for both Linux and Windows using cmake. GPU support works with the Python package as well as the CLI version. See [Installing R package with GPU support](#installing-r-package-with-gpu-support) for special instructions for R.
|
||||
|
||||
An up-to-date version of the CUDA toolkit is required.
|
||||
|
||||
From the command line on Linux starting from the xgboost directory:
|
||||
|
||||
```bash
|
||||
$ mkdir build
|
||||
$ cd build
|
||||
$ cmake .. -DUSE_CUDA=ON
|
||||
$ make -j
|
||||
```
|
||||
**Windows requirements** for GPU build: only Visual C++ 2015 or 2013 with CUDA v8.0 were fully tested. Either install Visual C++ 2015 Build Tools separately, or as a part of Visual Studio 2015. If you already have Visual Studio 2017, the Visual C++ 2015 Toolchain componenet has to be installed using the VS 2017 Installer. Likely, you would need to use the VS2015 x64 Native Tools command prompt to run the cmake commands given below. In some situations, however, things run just fine from MSYS2 bash command line.
|
||||
|
||||
On Windows, using cmake, see what options for Generators you have for cmake, and choose one with [arch] replaced by Win64:
|
||||
```bash
|
||||
cmake -help
|
||||
```
|
||||
Then run cmake as:
|
||||
```bash
|
||||
$ mkdir build
|
||||
$ cd build
|
||||
$ cmake .. -G"Visual Studio 14 2015 Win64" -DUSE_CUDA=ON
|
||||
```
|
||||
To speed up compilation, compute version specific to your GPU could be passed to cmake as, e.g., `-DGPU_COMPUTE_VER=50`.
|
||||
The above cmake configuration run will create an xgboost.sln solution file in the build directory. Build this solution in release mode as a x64 build, either from Visual studio or from command line:
|
||||
```
|
||||
cmake --build . --target xgboost --config Release
|
||||
```
|
||||
If build seems to use only a single process, you might try to append an option like ` -- /m:6` to the above command.
|
||||
|
||||
### Windows Binaries
|
||||
|
||||
After the build process successfully ends, you will find a `xgboost.dll` library file inside `./lib/` folder, copy this file to the the API package folder like `python-package/xgboost` if you are using *python* API. And you are good to follow the below instructions.
|
||||
|
||||
Unofficial windows binaries and instructions on how to use them are hosted on [Guido Tapia's blog](http://www.picnet.com.au/blogs/guido/post/2016/09/22/xgboost-windows-x64-binaries-for-download/)
|
||||
|
||||
### Building with Multi-GPU support
|
||||
Multi-GPU support requires the [NCCL](https://developer.nvidia.com/nccl) library. With NCCL installed, run cmake as:
|
||||
```bash
|
||||
cmake .. -DUSE_CUDA=ON -DUSE_NCCL=ON -DNCCL_ROOT="<NCCL_DIRECTORY>"
|
||||
export LD_LIBRARY_PATH=<NCCL_DIRECTORY>/lib:$LD_LIBRARY_PATH
|
||||
```
|
||||
One can also pass NCCL_ROOT as an environment variable, in which case, this takes precedence over the cmake variable NCCL_ROOT.
|
||||
|
||||
### Customized Building
|
||||
|
||||
The configuration of xgboost can be modified by ```config.mk```
|
||||
- modify configuration on various distributed filesystem such as HDFS/Amazon S3/...
|
||||
- First copy [make/config.mk](../make/config.mk) to the project root, on which
|
||||
any local modification will be ignored by git, then modify the according flags.
|
||||
|
||||
|
||||
|
||||
## Python Package Installation
|
||||
|
||||
The python package is located at [python-package](../python-package).
|
||||
There are several ways to install the package:
|
||||
|
||||
1. Install system-widely, which requires root permission
|
||||
|
||||
```bash
|
||||
cd python-package; sudo python setup.py install
|
||||
```
|
||||
|
||||
You will however need Python `distutils` module for this to
|
||||
work. It is often part of the core python package or it can be installed using your
|
||||
package manager, e.g. in Debian use
|
||||
|
||||
```bash
|
||||
sudo apt-get install python-setuptools
|
||||
```
|
||||
|
||||
*NOTE: If you recompiled xgboost, then you need to reinstall it again to
|
||||
make the new library take effect*
|
||||
|
||||
2. Only set the environment variable `PYTHONPATH` to tell python where to find
|
||||
the library. For example, assume we cloned `xgboost` on the home directory
|
||||
`~`. then we can added the following line in `~/.bashrc`.
|
||||
It is ***recommended for developers*** who may change the codes. The changes will be immediately reflected once you pulled the code and rebuild the project (no need to call ```setup``` again)
|
||||
|
||||
```bash
|
||||
export PYTHONPATH=~/xgboost/python-package
|
||||
```
|
||||
|
||||
3. Install only for the current user.
|
||||
|
||||
```bash
|
||||
cd python-package; python setup.py develop --user
|
||||
```
|
||||
|
||||
4. If you are installing the latest xgboost version which requires compilation, add MinGW to the system PATH:
|
||||
|
||||
```python
|
||||
import os
|
||||
os.environ['PATH'] = os.environ['PATH'] + ';C:\\Program Files\\mingw-w64\\x86_64-5.3.0-posix-seh-rt_v4-rev0\\mingw64\\bin'
|
||||
```
|
||||
|
||||
## R Package Installation
|
||||
|
||||
### Installing pre-packaged version
|
||||
|
||||
You can install xgboost from CRAN just like any other R package:
|
||||
|
||||
```r
|
||||
install.packages("xgboost")
|
||||
```
|
||||
|
||||
Or you can install it from our weekly updated drat repo:
|
||||
|
||||
```r
|
||||
install.packages("drat", repos="https://cran.rstudio.com")
|
||||
drat:::addRepo("dmlc")
|
||||
install.packages("xgboost", repos="http://dmlc.ml/drat/", type = "source")
|
||||
```
|
||||
|
||||
For OSX users, single threaded version will be installed. To install multi-threaded version,
|
||||
first follow [Building on OSX](#building-on-osx) to get the OpenMP enabled compiler, then:
|
||||
|
||||
- Set the `Makevars` file in highest piority for R.
|
||||
|
||||
The point is, there are three `Makevars` : `~/.R/Makevars`, `xgboost/R-package/src/Makevars`, and `/usr/local/Cellar/r/3.2.0/R.framework/Resources/etc/Makeconf` (the last one obtained by running `file.path(R.home("etc"), "Makeconf")` in R), and `SHLIB_OPENMP_CXXFLAGS` is not set by default!! After trying, it seems that the first one has highest piority (surprise!).
|
||||
|
||||
Then inside R, run
|
||||
|
||||
```R
|
||||
install.packages("drat", repos="https://cran.rstudio.com")
|
||||
drat:::addRepo("dmlc")
|
||||
install.packages("xgboost", repos="http://dmlc.ml/drat/", type = "source")
|
||||
```
|
||||
|
||||
### Installing the development version
|
||||
|
||||
Make sure you have installed git and a recent C++ compiler supporting C++11 (e.g., g++-4.8 or higher).
|
||||
On Windows, Rtools must be installed, and its bin directory has to be added to PATH during the installation.
|
||||
And see the previous subsection for an OSX tip.
|
||||
|
||||
Due to the use of git-submodules, `devtools::install_github` can no longer be used to install the latest version of R package.
|
||||
Thus, one has to run git to check out the code first:
|
||||
|
||||
```bash
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost
|
||||
git submodule init
|
||||
git submodule update
|
||||
cd R-package
|
||||
R CMD INSTALL .
|
||||
```
|
||||
|
||||
If the last line fails because of "R: command not found", it means that R was not set up to run from command line.
|
||||
In this case, just start R as you would normally do and run the following:
|
||||
|
||||
```r
|
||||
setwd('wherever/you/cloned/it/xgboost/R-package/')
|
||||
install.packages('.', repos = NULL, type="source")
|
||||
```
|
||||
|
||||
The package could also be built and installed with cmake (and Visual C++ 2015 on Windows) using instructions from the next section, but without GPU support (omit the `-DUSE_CUDA=ON` cmake parameter).
|
||||
|
||||
If all fails, try [building the shared library](#build-the-shared-library) to see whether a problem is specific to R package or not.
|
||||
|
||||
### Installing R package with GPU support
|
||||
|
||||
The procedure and requirements are similar as in [Building with GPU support](#building-with-gpu-support), so make sure to read it first.
|
||||
|
||||
On Linux, starting from the xgboost directory:
|
||||
|
||||
```bash
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -DUSE_CUDA=ON -DR_LIB=ON
|
||||
make install -j
|
||||
```
|
||||
When default target is used, an R package shared library would be built in the `build` area.
|
||||
The `install` target, in addition, assembles the package files with this shared library under `build/R-package`, and runs `R CMD INSTALL`.
|
||||
|
||||
On Windows, cmake with Visual C++ Build Tools (or Visual Studio) has to be used to build an R package with GPU support. Rtools must also be installed (perhaps, some other MinGW distributions with `gendef.exe` and `dlltool.exe` would work, but that was not tested).
|
||||
```bash
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -G"Visual Studio 14 2015 Win64" -DUSE_CUDA=ON -DR_LIB=ON
|
||||
cmake --build . --target install --config Release
|
||||
```
|
||||
When `--target xgboost` is used, an R package dll would be built under `build/Release`.
|
||||
The `--target install`, in addition, assembles the package files with this dll under `build/R-package`, and runs `R CMD INSTALL`.
|
||||
|
||||
If cmake can't find your R during the configuration step, you might provide the location of its executable to cmake like this: `-DLIBR_EXECUTABLE="C:/Program Files/R/R-3.4.1/bin/x64/R.exe"`.
|
||||
|
||||
If on Windows you get a "permission denied" error when trying to write to ...Program Files/R/... during the package installation, create a `.Rprofile` file in your personal home directory (if you don't already have one in there), and add a line to it which specifies the location of your R packages user library, like the following:
|
||||
```r
|
||||
.libPaths( unique(c("C:/Users/USERNAME/Documents/R/win-library/3.4", .libPaths())))
|
||||
```
|
||||
You might find the exact location by running `.libPaths()` in R GUI or RStudio.
|
||||
|
||||
## Trouble Shooting
|
||||
|
||||
1. **Compile failed after `git pull`**
|
||||
|
||||
Please first update the submodules, clean all and recompile:
|
||||
|
||||
```bash
|
||||
git submodule update && make clean_all && make -j4
|
||||
```
|
||||
|
||||
2. **Compile failed after `config.mk` is modified**
|
||||
|
||||
Need to clean all first:
|
||||
|
||||
```bash
|
||||
make clean_all && make -j4
|
||||
```
|
||||
|
||||
|
||||
3. **Makefile: dmlc-core/make/dmlc.mk: No such file or directory**
|
||||
|
||||
We need to recursively clone the submodule, you can do:
|
||||
|
||||
```bash
|
||||
git submodule init
|
||||
git submodule update
|
||||
```
|
||||
Alternatively, do another clone
|
||||
```bash
|
||||
git clone https://github.com/dmlc/xgboost --recursive
|
||||
```
|
||||
434
doc/build.rst
Normal file
434
doc/build.rst
Normal file
@ -0,0 +1,434 @@
|
||||
##################
|
||||
Installation Guide
|
||||
##################
|
||||
|
||||
.. note:: Pre-built binary wheel for Python
|
||||
|
||||
If you are planning to use Python on a Linux system, consider installing XGBoost from a pre-built binary wheel. The wheel is available from Python Package Index (PyPI). You may download and install it by running
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
# Ensure that you are downloading xgboost-{version}-py2.py3-none-manylinux1_x86_64.whl
|
||||
pip3 install xgboost
|
||||
|
||||
* This package will support GPU algorithms (`gpu_exact`, `gpu_hist`) on machines with NVIDIA GPUs.
|
||||
* Currently, PyPI has a binary wheel only for 64-bit Linux.
|
||||
|
||||
****************************
|
||||
Building XGBoost from source
|
||||
****************************
|
||||
This page gives instructions on how to build and install XGBoost from scratch on various systems. It consists of two steps:
|
||||
|
||||
1. First build the shared library from the C++ codes (``libxgboost.so`` for Linux/OSX and ``xgboost.dll`` for Windows).
|
||||
(For R-package installation, please directly refer to `R Package Installation`_.)
|
||||
2. Then install the language packages (e.g. Python Package).
|
||||
|
||||
.. note:: Use of Git submodules
|
||||
|
||||
XGBoost uses Git submodules to manage dependencies. So when you clone the repo, remember to specify ``--recursive`` option:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
|
||||
For windows users who use github tools, you can open the git shell and type the following command:
|
||||
|
||||
.. code-block:: batch
|
||||
|
||||
git submodule init
|
||||
git submodule update
|
||||
|
||||
Please refer to `Trouble Shooting`_ section first if you have any problem
|
||||
during installation. If the instructions do not work for you, please feel free
|
||||
to ask questions at `the user forum <https://discuss.xgboost.ai>`_.
|
||||
|
||||
**Contents**
|
||||
|
||||
* `Building the Shared Library`_
|
||||
|
||||
- `Building on Ubuntu/Debian`_
|
||||
- `Building on OSX`_
|
||||
- `Building on Windows`_
|
||||
- `Building with GPU support`_
|
||||
- `Customized Building`_
|
||||
|
||||
* `Python Package Installation`_
|
||||
* `R Package Installation`_
|
||||
* `Trouble Shooting`_
|
||||
|
||||
***************************
|
||||
Building the Shared Library
|
||||
***************************
|
||||
|
||||
Our goal is to build the shared library:
|
||||
|
||||
- On Linux/OSX the target library is ``libxgboost.so``
|
||||
- On Windows the target library is ``xgboost.dll``
|
||||
|
||||
The minimal building requirement is
|
||||
|
||||
- A recent C++ compiler supporting C++11 (g++-4.8 or higher)
|
||||
|
||||
We can edit ``make/config.mk`` to change the compile options, and then build by
|
||||
``make``. If everything goes well, we can go to the specific language installation section.
|
||||
|
||||
Building on Ubuntu/Debian
|
||||
=========================
|
||||
|
||||
On Ubuntu, one builds XGBoost by running
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost; make -j4
|
||||
|
||||
Building on OSX
|
||||
===============
|
||||
|
||||
Install with pip: simple method
|
||||
--------------------------------
|
||||
|
||||
First, make sure you obtained ``gcc-5`` (newer version does not work with this method yet). Note: installation of ``gcc`` can take a while (~ 30 minutes).
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
brew install gcc@5
|
||||
|
||||
Then install XGBoost with ``pip``:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
pip3 install xgboost
|
||||
|
||||
You might need to run the command with ``sudo`` if you run into permission errors.
|
||||
|
||||
Build from the source code - advanced method
|
||||
--------------------------------------------
|
||||
|
||||
First, obtain ``gcc-7`` with homebrew (https://brew.sh/) if you want multi-threaded version. Clang is okay if multithreading is not required. Note: installation of ``gcc`` can take a while (~ 30 minutes).
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
brew install gcc@7
|
||||
|
||||
Now, clone the repository:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost; cp make/config.mk ./config.mk
|
||||
|
||||
Open ``config.mk`` and uncomment these two lines:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
export CC = gcc
|
||||
export CXX = g++
|
||||
|
||||
and replace these two lines as follows: (specify the GCC version)
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
export CC = gcc-7
|
||||
export CXX = g++-7
|
||||
|
||||
Now, you may build XGBoost using the following command:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make -j4
|
||||
|
||||
You may now continue to `Python Package Installation`_.
|
||||
|
||||
Building on Windows
|
||||
===================
|
||||
You need to first clone the XGBoost repo with ``--recursive`` option, to clone the submodules.
|
||||
We recommend you use `Git for Windows <https://git-for-windows.github.io/>`_, as it comes with a standard Bash shell. This will highly ease the installation process.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git submodule init
|
||||
git submodule update
|
||||
|
||||
XGBoost support compilation with Microsoft Visual Studio and MinGW.
|
||||
|
||||
Compile XGBoost using MinGW
|
||||
---------------------------
|
||||
After installing `Git for Windows <https://git-for-windows.github.io/>`_, you should have a shortcut named ``Git Bash``. You should run all subsequent steps in ``Git Bash``.
|
||||
|
||||
In MinGW, ``make`` command comes with the name ``mingw32-make``. You can add the following line into the ``.bashrc`` file:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
alias make='mingw32-make'
|
||||
|
||||
(On 64-bit Windows, you should get `MinGW64 <https://sourceforge.net/projects/mingw-w64/>`_ instead.) Make sure
|
||||
that the path to MinGW is in the system PATH.
|
||||
|
||||
To build with MinGW, type:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cp make/mingw64.mk config.mk; make -j4
|
||||
|
||||
Compile XGBoost with Microsoft Visual Studio
|
||||
--------------------------------------------
|
||||
To build with Visual Studio, we will need CMake. Make sure to install a recent version of CMake. Then run the following from the root of the XGBoost directory:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -G"Visual Studio 12 2013 Win64"
|
||||
|
||||
This specifies an out of source build using the MSVC 12 64 bit generator. Open the ``.sln`` file in the build directory and build with Visual Studio. To use the Python module you can copy ``xgboost.dll`` into ``python-package/xgboost``.
|
||||
|
||||
After the build process successfully ends, you will find a ``xgboost.dll`` library file inside ``./lib/`` folder, copy this file to the the API package folder like ``python-package/xgboost`` if you are using Python API.
|
||||
|
||||
Unofficial windows binaries and instructions on how to use them are hosted on `Guido Tapia's blog <http://www.picnet.com.au/blogs/guido/post/2016/09/22/xgboost-windows-x64-binaries-for-download/>`_.
|
||||
|
||||
Building with GPU support
|
||||
=========================
|
||||
XGBoost can be built with GPU support for both Linux and Windows using CMake. GPU support works with the Python package as well as the CLI version. See `Installing R package with GPU support`_ for special instructions for R.
|
||||
|
||||
An up-to-date version of the CUDA toolkit is required.
|
||||
|
||||
From the command line on Linux starting from the xgboost directory:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -DUSE_CUDA=ON
|
||||
make -j
|
||||
|
||||
.. note:: Windows requirements for GPU build
|
||||
|
||||
Only Visual C++ 2015 or 2013 with CUDA v8.0 were fully tested. Either install Visual C++ 2015 Build Tools separately, or as a part of Visual Studio 2015. If you already have Visual Studio 2017, the Visual C++ 2015 Toolchain componenet has to be installed using the VS 2017 Installer. Likely, you would need to use the VS2015 x64 Native Tools command prompt to run the cmake commands given below. In some situations, however, things run just fine from MSYS2 bash command line.
|
||||
|
||||
On Windows, see what options for generators you have for CMake, and choose one with ``[arch]`` replaced with Win64:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cmake -help
|
||||
|
||||
Then run CMake as follows:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -G"Visual Studio 14 2015 Win64" -DUSE_CUDA=ON
|
||||
|
||||
.. note:: Visual Studio 2017 Win64 Generator may not work
|
||||
|
||||
Choosing the Visual Studio 2017 generator may cause compilation failure. When it happens, specify the 2015 compiler by adding the ``-T`` option:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make .. -G"Visual Studio 15 2017 Win64" -T v140,cuda=8.0 -DR_LIB=ON -DUSE_CUDA=ON
|
||||
|
||||
To speed up compilation, the compute version specific to your GPU could be passed to cmake as, e.g., ``-DGPU_COMPUTE_VER=50``.
|
||||
The above cmake configuration run will create an ``xgboost.sln`` solution file in the build directory. Build this solution in release mode as a x64 build, either from Visual studio or from command line:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cmake --build . --target xgboost --config Release
|
||||
|
||||
To speed up compilation, run multiple jobs in parallel by appending option ``-- /MP``.
|
||||
|
||||
Customized Building
|
||||
===================
|
||||
|
||||
The configuration file ``config.mk`` modifies several compilation flags:
|
||||
- Whether to enable support for various distributed filesystems such as HDFS and Amazon S3
|
||||
- Which compiler to use
|
||||
- And some more
|
||||
|
||||
To customize, first copy ``make/config.mk`` to the project root and then modify the copy.
|
||||
|
||||
Python Package Installation
|
||||
===========================
|
||||
|
||||
The python package is located at ``python-package/``.
|
||||
There are several ways to install the package:
|
||||
|
||||
1. Install system-wide, which requires root permission:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cd python-package; sudo python setup.py install
|
||||
|
||||
You will however need Python ``distutils`` module for this to
|
||||
work. It is often part of the core python package or it can be installed using your
|
||||
package manager, e.g. in Debian use
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
sudo apt-get install python-setuptools
|
||||
|
||||
.. note:: Re-compiling XGBoost
|
||||
|
||||
If you recompiled XGBoost, then you need to reinstall it again to make the new library take effect.
|
||||
|
||||
2. Only set the environment variable ``PYTHONPATH`` to tell python where to find
|
||||
the library. For example, assume we cloned `xgboost` on the home directory
|
||||
`~`. then we can added the following line in `~/.bashrc`.
|
||||
This option is **recommended for developers** who change the code frequently. The changes will be immediately reflected once you pulled the code and rebuild the project (no need to call ``setup`` again)
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
export PYTHONPATH=~/xgboost/python-package
|
||||
|
||||
3. Install only for the current user.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cd python-package; python setup.py develop --user
|
||||
|
||||
4. If you are installing the latest XGBoost version which requires compilation, add MinGW to the system PATH:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
import os
|
||||
os.environ['PATH'] = os.environ['PATH'] + ';C:\\Program Files\\mingw-w64\\x86_64-5.3.0-posix-seh-rt_v4-rev0\\mingw64\\bin'
|
||||
|
||||
R Package Installation
|
||||
======================
|
||||
|
||||
Installing pre-packaged version
|
||||
-------------------------------
|
||||
|
||||
You can install xgboost from CRAN just like any other R package:
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
install.packages("xgboost")
|
||||
|
||||
Or you can install it from our weekly updated drat repo:
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
install.packages("drat", repos="https://cran.rstudio.com")
|
||||
drat:::addRepo("dmlc")
|
||||
install.packages("xgboost", repos="http://dmlc.ml/drat/", type = "source")
|
||||
|
||||
For OSX users, single threaded version will be installed. To install multi-threaded version,
|
||||
first follow `Building on OSX`_ to get the OpenMP enabled compiler. Then
|
||||
|
||||
- Set the ``Makevars`` file in highest piority for R.
|
||||
|
||||
The point is, there are three ``Makevars`` : ``~/.R/Makevars``, ``xgboost/R-package/src/Makevars``, and ``/usr/local/Cellar/r/3.2.0/R.framework/Resources/etc/Makeconf`` (the last one obtained by running ``file.path(R.home("etc"), "Makeconf")`` in R), and ``SHLIB_OPENMP_CXXFLAGS`` is not set by default!! After trying, it seems that the first one has highest piority (surprise!).
|
||||
|
||||
Then inside R, run
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
install.packages("drat", repos="https://cran.rstudio.com")
|
||||
drat:::addRepo("dmlc")
|
||||
install.packages("xgboost", repos="http://dmlc.ml/drat/", type = "source")
|
||||
|
||||
Installing the development version
|
||||
----------------------------------
|
||||
|
||||
Make sure you have installed git and a recent C++ compiler supporting C++11 (e.g., g++-4.8 or higher).
|
||||
On Windows, Rtools must be installed, and its bin directory has to be added to PATH during the installation.
|
||||
And see the previous subsection for an OSX tip.
|
||||
|
||||
Due to the use of git-submodules, ``devtools::install_github`` can no longer be used to install the latest version of R package.
|
||||
Thus, one has to run git to check out the code first:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost
|
||||
git submodule init
|
||||
git submodule update
|
||||
cd R-package
|
||||
R CMD INSTALL .
|
||||
|
||||
If the last line fails because of the error ``R: command not found``, it means that R was not set up to run from command line.
|
||||
In this case, just start R as you would normally do and run the following:
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
setwd('wherever/you/cloned/it/xgboost/R-package/')
|
||||
install.packages('.', repos = NULL, type="source")
|
||||
|
||||
The package could also be built and installed with cmake (and Visual C++ 2015 on Windows) using instructions from the next section, but without GPU support (omit the ``-DUSE_CUDA=ON`` cmake parameter).
|
||||
|
||||
If all fails, try `Building the shared library`_ to see whether a problem is specific to R package or not.
|
||||
|
||||
Installing R package with GPU support
|
||||
-------------------------------------
|
||||
|
||||
The procedure and requirements are similar as in `Building with GPU support`_, so make sure to read it first.
|
||||
|
||||
On Linux, starting from the XGBoost directory type:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -DUSE_CUDA=ON -DR_LIB=ON
|
||||
make install -j
|
||||
|
||||
When default target is used, an R package shared library would be built in the ``build`` area.
|
||||
The ``install`` target, in addition, assembles the package files with this shared library under ``build/R-package``, and runs ``R CMD INSTALL``.
|
||||
|
||||
On Windows, cmake with Visual C++ Build Tools (or Visual Studio) has to be used to build an R package with GPU support. Rtools must also be installed (perhaps, some other MinGW distributions with ``gendef.exe`` and ``dlltool.exe`` would work, but that was not tested).
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mkdir build
|
||||
cd build
|
||||
cmake .. -G"Visual Studio 14 2015 Win64" -DUSE_CUDA=ON -DR_LIB=ON
|
||||
cmake --build . --target install --config Release
|
||||
|
||||
When ``--target xgboost`` is used, an R package dll would be built under ``build/Release``.
|
||||
The ``--target install``, in addition, assembles the package files with this dll under ``build/R-package``, and runs ``R CMD INSTALL``.
|
||||
|
||||
If cmake can't find your R during the configuration step, you might provide the location of its executable to cmake like this: ``-DLIBR_EXECUTABLE="C:/Program Files/R/R-3.4.1/bin/x64/R.exe"``.
|
||||
|
||||
If on Windows you get a "permission denied" error when trying to write to ...Program Files/R/... during the package installation, create a ``.Rprofile`` file in your personal home directory (if you don't already have one in there), and add a line to it which specifies the location of your R packages user library, like the following:
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
.libPaths( unique(c("C:/Users/USERNAME/Documents/R/win-library/3.4", .libPaths())))
|
||||
|
||||
You might find the exact location by running ``.libPaths()`` in R GUI or RStudio.
|
||||
|
||||
Trouble Shooting
|
||||
================
|
||||
|
||||
1. Compile failed after ``git pull``
|
||||
|
||||
Please first update the submodules, clean all and recompile:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git submodule update && make clean_all && make -j4
|
||||
|
||||
2. Compile failed after ``config.mk`` is modified
|
||||
|
||||
Need to clean all first:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make clean_all && make -j4
|
||||
|
||||
3. ``Makefile: dmlc-core/make/dmlc.mk: No such file or directory``
|
||||
|
||||
We need to recursively clone the submodule:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git submodule init
|
||||
git submodule update
|
||||
|
||||
Alternatively, do another clone
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone https://github.com/dmlc/xgboost --recursive
|
||||
|
||||
5
doc/cli.rst
Normal file
5
doc/cli.rst
Normal file
@ -0,0 +1,5 @@
|
||||
############################
|
||||
XGBoost Command Line version
|
||||
############################
|
||||
|
||||
See `XGBoost Command Line walkthrough <https://github.com/dmlc/xgboost/blob/master/demo/binary_classification/README.md>`_.
|
||||
@ -1,3 +0,0 @@
|
||||
# XGBoost Command Line version
|
||||
|
||||
See [XGBoost Command Line walkthrough](https://github.com/dmlc/xgboost/blob/master/demo/binary_classification/README.md)
|
||||
88
doc/conf.py
88
doc/conf.py
@ -11,9 +11,12 @@
|
||||
#
|
||||
# All configuration values have a default; values that are commented out
|
||||
# serve to show the default.
|
||||
from subprocess import call
|
||||
from recommonmark.parser import CommonMarkParser
|
||||
import sys
|
||||
import os, subprocess
|
||||
import shlex
|
||||
import guzzle_sphinx_theme
|
||||
# If extensions (or modules to document with autodoc) are in another directory,
|
||||
# add these directories to sys.path here. If the directory is relative to the
|
||||
# documentation root, use os.path.abspath to make it absolute, like shown here.
|
||||
@ -22,13 +25,11 @@ libpath = os.path.join(curr_path, '../python-package/')
|
||||
sys.path.insert(0, libpath)
|
||||
sys.path.insert(0, curr_path)
|
||||
|
||||
from sphinx_util import MarkdownParser, AutoStructify
|
||||
|
||||
# -- mock out modules
|
||||
import mock
|
||||
MOCK_MODULES = ['numpy', 'scipy', 'scipy.sparse', 'sklearn', 'matplotlib', 'pandas', 'graphviz']
|
||||
for mod_name in MOCK_MODULES:
|
||||
sys.modules[mod_name] = mock.Mock()
|
||||
sys.modules[mod_name] = mock.Mock()
|
||||
|
||||
# -- General configuration ------------------------------------------------
|
||||
|
||||
@ -38,11 +39,6 @@ author = u'%s developers' % project
|
||||
copyright = u'2016, %s' % author
|
||||
github_doc_root = 'https://github.com/dmlc/xgboost/tree/master/doc/'
|
||||
|
||||
# add markdown parser
|
||||
MarkdownParser.github_doc_root = github_doc_root
|
||||
source_parsers = {
|
||||
'.md': MarkdownParser,
|
||||
}
|
||||
os.environ['XGBOOST_BUILD_DOC'] = '1'
|
||||
# Version information.
|
||||
import xgboost
|
||||
@ -55,14 +51,23 @@ extensions = [
|
||||
'sphinx.ext.autodoc',
|
||||
'sphinx.ext.napoleon',
|
||||
'sphinx.ext.mathjax',
|
||||
'sphinx.ext.intersphinx',
|
||||
'breathe'
|
||||
]
|
||||
|
||||
# Breathe extension variables
|
||||
breathe_projects = {"xgboost": "doxyxml/"}
|
||||
breathe_default_project = "xgboost"
|
||||
|
||||
# Add any paths that contain templates here, relative to this directory.
|
||||
templates_path = ['_templates']
|
||||
|
||||
source_parsers = {
|
||||
'.md': CommonMarkParser,
|
||||
}
|
||||
|
||||
# The suffix(es) of source filenames.
|
||||
# You can specify multiple suffix as a list of string:
|
||||
# source_suffix = ['.rst', '.md']
|
||||
source_suffix = ['.rst', '.md']
|
||||
|
||||
# The encoding of source files.
|
||||
@ -119,11 +124,23 @@ todo_include_todos = False
|
||||
|
||||
# -- Options for HTML output ----------------------------------------------
|
||||
|
||||
html_theme_path = ['_static']
|
||||
# The theme to use for HTML and HTML Help pages. See the documentation for
|
||||
# a list of builtin themes.
|
||||
# html_theme = 'alabaster'
|
||||
html_theme = 'xgboost-theme'
|
||||
html_theme_path = guzzle_sphinx_theme.html_theme_path()
|
||||
html_theme = 'guzzle_sphinx_theme'
|
||||
|
||||
# Register the theme as an extension to generate a sitemap.xml
|
||||
extensions.append("guzzle_sphinx_theme")
|
||||
|
||||
# Guzzle theme options (see theme.conf for more information)
|
||||
html_theme_options = {
|
||||
# Set the name of the project to appear in the sidebar
|
||||
"project_nav_name": "XGBoost"
|
||||
}
|
||||
|
||||
html_sidebars = {
|
||||
'**': ['logo-text.html', 'globaltoc.html', 'searchbox.html']
|
||||
}
|
||||
|
||||
# Add any paths that contain custom static files (such as style sheets) here,
|
||||
# relative to this directory. They are copied after the builtin static files,
|
||||
@ -145,38 +162,27 @@ latex_documents = [
|
||||
author, 'manual'),
|
||||
]
|
||||
|
||||
intersphinx_mapping = {'python': ('https://docs.python.org/3.6', None),
|
||||
'numpy': ('http://docs.scipy.org/doc/numpy/', None),
|
||||
'scipy': ('http://docs.scipy.org/doc/scipy/reference/', None),
|
||||
'pandas': ('http://pandas-docs.github.io/pandas-docs-travis/', None),
|
||||
'sklearn': ('http://scikit-learn.org/stable', None)}
|
||||
|
||||
# hook for doxygen
|
||||
def run_doxygen(folder):
|
||||
"""Run the doxygen make command in the designated folder."""
|
||||
try:
|
||||
retcode = subprocess.call("cd %s; make doxygen" % folder, shell=True)
|
||||
if retcode < 0:
|
||||
sys.stderr.write("doxygen terminated by signal %s" % (-retcode))
|
||||
except OSError as e:
|
||||
sys.stderr.write("doxygen execution failed: %s" % e)
|
||||
"""Run the doxygen make command in the designated folder."""
|
||||
try:
|
||||
retcode = subprocess.call("cd %s; make doxygen" % folder, shell=True)
|
||||
if retcode < 0:
|
||||
sys.stderr.write("doxygen terminated by signal %s" % (-retcode))
|
||||
except OSError as e:
|
||||
sys.stderr.write("doxygen execution failed: %s" % e)
|
||||
|
||||
def generate_doxygen_xml(app):
|
||||
"""Run the doxygen make commands if we're on the ReadTheDocs server"""
|
||||
read_the_docs_build = os.environ.get('READTHEDOCS', None) == 'True'
|
||||
if read_the_docs_build:
|
||||
run_doxygen('..')
|
||||
"""Run the doxygen make commands if we're on the ReadTheDocs server"""
|
||||
read_the_docs_build = os.environ.get('READTHEDOCS', None) == 'True'
|
||||
if read_the_docs_build:
|
||||
run_doxygen('..')
|
||||
|
||||
def setup(app):
|
||||
# Add hook for building doxygen xml when needed
|
||||
# no c++ API for now
|
||||
# app.connect("builder-inited", generate_doxygen_xml)
|
||||
|
||||
# urlretrieve got moved in Python 3.x
|
||||
try:
|
||||
from urllib import urlretrieve
|
||||
except ImportError:
|
||||
from urllib.request import urlretrieve
|
||||
urlretrieve('https://code.jquery.com/jquery-2.2.4.min.js',
|
||||
'_static/jquery.js')
|
||||
app.add_config_value('recommonmark_config', {
|
||||
'url_resolver': lambda url: github_doc_root + url,
|
||||
'enable_eval_rst': True,
|
||||
}, True,
|
||||
)
|
||||
app.add_transform(AutoStructify)
|
||||
app.add_javascript('jquery.js')
|
||||
app.add_stylesheet('custom.css')
|
||||
|
||||
215
doc/contribute.rst
Normal file
215
doc/contribute.rst
Normal file
@ -0,0 +1,215 @@
|
||||
#####################
|
||||
Contribute to XGBoost
|
||||
#####################
|
||||
XGBoost has been developed and used by a group of active community members.
|
||||
Everyone is more than welcome to contribute. It is a way to make the project better and more accessible to more users.
|
||||
|
||||
- Please add your name to `CONTRIBUTORS.md <https://github.com/dmlc/xgboost/blob/master/CONTRIBUTORS.md>`_ after your patch has been merged.
|
||||
- Please also update `NEWS.md <https://github.com/dmlc/xgboost/blob/master/NEWS.md>`_ to add note on your changes to the API or XGBoost documentation.
|
||||
|
||||
**Guidelines**
|
||||
|
||||
* `Submit Pull Request`_
|
||||
* `Git Workflow Howtos`_
|
||||
|
||||
- `How to resolve conflict with master`_
|
||||
- `How to combine multiple commits into one`_
|
||||
- `What is the consequence of force push`_
|
||||
|
||||
* `Documents`_
|
||||
* `Testcases`_
|
||||
* `Examples`_
|
||||
* `Core Library`_
|
||||
* `Python Package`_
|
||||
* `R Package`_
|
||||
|
||||
*******************
|
||||
Submit Pull Request
|
||||
*******************
|
||||
|
||||
* Before submit, please rebase your code on the most recent version of master, you can do it by
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git remote add upstream https://github.com/dmlc/xgboost
|
||||
git fetch upstream
|
||||
git rebase upstream/master
|
||||
|
||||
* If you have multiple small commits,
|
||||
it might be good to merge them together(use git rebase then squash) into more meaningful groups.
|
||||
* Send the pull request!
|
||||
|
||||
- Fix the problems reported by automatic checks
|
||||
- If you are contributing a new module, consider add a testcase in `tests <https://github.com/dmlc/xgboost/tree/master/tests>`_.
|
||||
|
||||
*******************
|
||||
Git Workflow Howtos
|
||||
*******************
|
||||
|
||||
How to resolve conflict with master
|
||||
===================================
|
||||
- First rebase to most recent master
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
# The first two steps can be skipped after you do it once.
|
||||
git remote add upstream https://github.com/dmlc/xgboost
|
||||
git fetch upstream
|
||||
git rebase upstream/master
|
||||
|
||||
- The git may show some conflicts it cannot merge, say ``conflicted.py``.
|
||||
|
||||
- Manually modify the file to resolve the conflict.
|
||||
- After you resolved the conflict, mark it as resolved by
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git add conflicted.py
|
||||
|
||||
- Then you can continue rebase by
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git rebase --continue
|
||||
|
||||
- Finally push to your fork, you may need to force push here.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git push --force
|
||||
|
||||
How to combine multiple commits into one
|
||||
========================================
|
||||
Sometimes we want to combine multiple commits, especially when later commits are only fixes to previous ones,
|
||||
to create a PR with set of meaningful commits. You can do it by following steps.
|
||||
|
||||
- Before doing so, configure the default editor of git if you haven't done so before.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git config core.editor the-editor-you-like
|
||||
|
||||
- Assume we want to merge last 3 commits, type the following commands
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git rebase -i HEAD~3
|
||||
|
||||
- It will pop up an text editor. Set the first commit as ``pick``, and change later ones to ``squash``.
|
||||
- After you saved the file, it will pop up another text editor to ask you modify the combined commit message.
|
||||
- Push the changes to your fork, you need to force push.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git push --force
|
||||
|
||||
What is the consequence of force push
|
||||
=====================================
|
||||
The previous two tips requires force push, this is because we altered the path of the commits.
|
||||
It is fine to force push to your own fork, as long as the commits changed are only yours.
|
||||
|
||||
*********
|
||||
Documents
|
||||
*********
|
||||
* Documentation is built using sphinx.
|
||||
* Each document is written in `reStructuredText <http://www.sphinx-doc.org/en/master/usage/restructuredtext/basics.html>`_.
|
||||
* You can build document locally to see the effect.
|
||||
|
||||
*********
|
||||
Testcases
|
||||
*********
|
||||
* All the testcases are in `tests <https://github.com/dmlc/xgboost/tree/master/tests>`_.
|
||||
* We use python nose for python test cases.
|
||||
|
||||
********
|
||||
Examples
|
||||
********
|
||||
* Usecases and examples will be in `demo <https://github.com/dmlc/xgboost/tree/master/demo>`_.
|
||||
* We are super excited to hear about your story, if you have blogposts,
|
||||
tutorials code solutions using XGBoost, please tell us and we will add
|
||||
a link in the example pages.
|
||||
|
||||
************
|
||||
Core Library
|
||||
************
|
||||
- Follow `Google style for C++ <https://google.github.io/styleguide/cppguide.html>`_.
|
||||
- Use C++11 features such as smart pointers, braced initializers, lambda functions, and ``std::thread``.
|
||||
- We use Doxygen to document all the interface code.
|
||||
- You can reproduce the linter checks by running ``make lint``
|
||||
|
||||
**************
|
||||
Python Package
|
||||
**************
|
||||
- Always add docstring to the new functions in numpydoc format.
|
||||
- You can reproduce the linter checks by typing ``make lint``
|
||||
|
||||
*********
|
||||
R Package
|
||||
*********
|
||||
|
||||
Code Style
|
||||
==========
|
||||
- We follow Google's C++ Style guide for C++ code.
|
||||
|
||||
- This is mainly to be consistent with the rest of the project.
|
||||
- Another reason is we will be able to check style automatically with a linter.
|
||||
|
||||
- You can check the style of the code by typing the following command at root folder.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make rcpplint
|
||||
|
||||
- When needed, you can disable the linter warning of certain line with ```// NOLINT(*)``` comments.
|
||||
- We use `roxygen <https://cran.r-project.org/web/packages/roxygen2/vignettes/roxygen2.html>`_ for documenting the R package.
|
||||
|
||||
Rmarkdown Vignettes
|
||||
===================
|
||||
Rmarkdown vignettes are placed in `R-package/vignettes <https://github.com/dmlc/xgboost/tree/master/R-package/vignettes>`_.
|
||||
These Rmarkdown files are not compiled. We host the compiled version on `doc/R-package <https://github.com/dmlc/xgboost/tree/master/doc/R-package>`_.
|
||||
|
||||
The following steps are followed to add a new Rmarkdown vignettes:
|
||||
|
||||
- Add the original rmarkdown to ``R-package/vignettes``.
|
||||
- Modify ``doc/R-package/Makefile`` to add the markdown files to be build.
|
||||
- Clone the `dmlc/web-data <https://github.com/dmlc/web-data>`_ repo to folder ``doc``.
|
||||
- Now type the following command on ``doc/R-package``:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make the-markdown-to-make.md
|
||||
|
||||
- This will generate the markdown, as well as the figures in ``doc/web-data/xgboost/knitr``.
|
||||
- Modify the ``doc/R-package/index.md`` to point to the generated markdown.
|
||||
- Add the generated figure to the ``dmlc/web-data`` repo.
|
||||
|
||||
- If you already cloned the repo to doc, this means ``git add``
|
||||
|
||||
- Create PR for both the markdown and ``dmlc/web-data``.
|
||||
- You can also build the document locally by typing the following command at the ``doc`` directory:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
make html
|
||||
|
||||
The reason we do this is to avoid exploded repo size due to generated images.
|
||||
|
||||
R package versioning
|
||||
====================
|
||||
Since version 0.6.4.3, we have adopted a versioning system that uses x.y.z (or ``core_major.core_minor.cran_release``)
|
||||
format for CRAN releases and an x.y.z.p (or ``core_major.core_minor.cran_release.patch``) format for development patch versions.
|
||||
This approach is similar to the one described in Yihui Xie's
|
||||
`blog post on R Package Versioning <https://yihui.name/en/2013/06/r-package-versioning/>`_,
|
||||
except we need an additional field to accomodate the x.y core library version.
|
||||
|
||||
Each new CRAN release bumps up the 3rd field, while developments in-between CRAN releases
|
||||
would be marked by an additional 4th field on the top of an existing CRAN release version.
|
||||
Some additional consideration is needed when the core library version changes.
|
||||
E.g., after the core changes from 0.6 to 0.7, the R package development version would become 0.7.0.1, working towards
|
||||
a 0.7.1 CRAN release. The 0.7.0 would not be released to CRAN, unless it would require almost no additional development.
|
||||
|
||||
Registering native routines in R
|
||||
================================
|
||||
According to `R extension manual <https://cran.r-project.org/doc/manuals/r-release/R-exts.html#Registering-native-routines>`_,
|
||||
it is good practice to register native routines and to disable symbol search. When any changes or additions are made to the
|
||||
C++ interface of the R package, please make corresponding changes in ``src/init.c`` as well.
|
||||
@ -1,46 +1,50 @@
|
||||
##########################
|
||||
Frequently Asked Questions
|
||||
========================
|
||||
This document contains frequently asked questions about xgboost.
|
||||
##########################
|
||||
|
||||
This document contains frequently asked questions about XGBoost.
|
||||
|
||||
**********************
|
||||
How to tune parameters
|
||||
----------------------
|
||||
See [Parameter Tunning Guide](how_to/param_tuning.md)
|
||||
**********************
|
||||
See :doc:`Parameter Tunning Guide </tutorials/param_tuning>`.
|
||||
|
||||
************************
|
||||
Description on the model
|
||||
------------------------
|
||||
See [Introduction to Boosted Trees](model.md)
|
||||
|
||||
************************
|
||||
See :doc:`Introduction to Boosted Trees </tutorials/model>`.
|
||||
|
||||
********************
|
||||
I have a big dataset
|
||||
--------------------
|
||||
XGBoost is designed to be memory efficient. Usually it can handle problems as long as the data fit into your memory
|
||||
(This usually means millions of instances).
|
||||
If you are running out of memory, checkout [external memory version](how_to/external_memory.md) or
|
||||
[distributed version](../demo/distributed-training) of xgboost.
|
||||
********************
|
||||
XGBoost is designed to be memory efficient. Usually it can handle problems as long as the data fit into your memory.
|
||||
(This usually means millions of instances)
|
||||
If you are running out of memory, checkout :doc:`external memory version </tutorials/external_memory>` or
|
||||
:doc:`distributed version </tutorials/aws_yarn>` of XGBoost.
|
||||
|
||||
|
||||
Running xgboost on Platform X (Hadoop/Yarn, Mesos)
|
||||
--------------------------------------------------
|
||||
**************************************************
|
||||
Running XGBoost on Platform X (Hadoop/Yarn, Mesos)
|
||||
**************************************************
|
||||
The distributed version of XGBoost is designed to be portable to various environment.
|
||||
Distributed XGBoost can be ported to any platform that supports [rabit](https://github.com/dmlc/rabit).
|
||||
You can directly run xgboost on Yarn. In theory Mesos and other resource allocation engines can be easily supported as well.
|
||||
Distributed XGBoost can be ported to any platform that supports `rabit <https://github.com/dmlc/rabit>`_.
|
||||
You can directly run XGBoost on Yarn. In theory Mesos and other resource allocation engines can be easily supported as well.
|
||||
|
||||
|
||||
Why not implement distributed xgboost on top of X (Spark, Hadoop)
|
||||
-----------------------------------------------------------------
|
||||
*****************************************************************
|
||||
Why not implement distributed XGBoost on top of X (Spark, Hadoop)
|
||||
*****************************************************************
|
||||
The first fact we need to know is going distributed does not necessarily solve all the problems.
|
||||
Instead, it creates more problems such as more communication overhead and fault tolerance.
|
||||
The ultimate question will still come back to how to push the limit of each computation node
|
||||
and use less resources to complete the task (thus with less communication and chance of failure).
|
||||
|
||||
To achieve these, we decide to reuse the optimizations in the single node xgboost and build distributed version on top of it.
|
||||
To achieve these, we decide to reuse the optimizations in the single node XGBoost and build distributed version on top of it.
|
||||
The demand of communication in machine learning is rather simple, in the sense that we can depend on a limited set of API (in our case rabit).
|
||||
Such design allows us to reuse most of the code, while being portable to major platforms such as Hadoop/Yarn, MPI, SGE.
|
||||
Most importantly, it pushes the limit of the computation resources we can use.
|
||||
|
||||
|
||||
*****************************************
|
||||
How can I port the model to my own system
|
||||
-----------------------------------------
|
||||
*****************************************
|
||||
The model and data format of XGBoost is exchangeable,
|
||||
which means the model trained by one language can be loaded in another.
|
||||
This means you can train the model using R, while running prediction using
|
||||
@ -48,26 +52,26 @@ Java or C++, which are more common in production systems.
|
||||
You can also train the model using distributed versions,
|
||||
and load them in from Python to do some interactive analysis.
|
||||
|
||||
|
||||
*************************
|
||||
Do you support LambdaMART
|
||||
-------------------------
|
||||
Yes, xgboost implements LambdaMART. Checkout the objective section in [parameters](parameter.md)
|
||||
|
||||
*************************
|
||||
Yes, XGBoost implements LambdaMART. Checkout the objective section in :doc:`parameters </parameter>`.
|
||||
|
||||
******************************
|
||||
How to deal with Missing Value
|
||||
------------------------------
|
||||
xgboost supports missing value by default.
|
||||
******************************
|
||||
XGBoost supports missing value by default.
|
||||
In tree algorithms, branch directions for missing values are learned during training.
|
||||
Note that the gblinear booster treats missing values as zeros.
|
||||
|
||||
|
||||
**************************************
|
||||
Slightly different result between runs
|
||||
--------------------------------------
|
||||
**************************************
|
||||
This could happen, due to non-determinism in floating point summation order and multi-threading.
|
||||
Though the general accuracy will usually remain the same.
|
||||
|
||||
|
||||
**********************************************************
|
||||
Why do I see different results with sparse and dense data?
|
||||
--------------------------------------------------------
|
||||
**********************************************************
|
||||
"Sparse" elements are treated as if they were "missing" by the tree booster, and as zeros by the linear booster.
|
||||
For tree models, it is important to use consistent data formats during training and scoring.
|
||||
For tree models, it is important to use consistent data formats during training and scoring.
|
||||
94
doc/get_started.rst
Normal file
94
doc/get_started.rst
Normal file
@ -0,0 +1,94 @@
|
||||
########################
|
||||
Get Started with XGBoost
|
||||
########################
|
||||
|
||||
This is a quick start tutorial showing snippets for you to quickly try out XGBoost
|
||||
on the demo dataset on a binary classification task.
|
||||
|
||||
********************************
|
||||
Links to Other Helpful Resources
|
||||
********************************
|
||||
- See :doc:`Installation Guide </build>` on how to install XGBoost.
|
||||
- See :doc:`Text Input Format </tutorials/input_format>` on using text format for specifying training/testing data.
|
||||
- See :doc:`Tutorials </tutorials/index>` for tips and tutorials.
|
||||
- See `Learning to use XGBoost by Examples <https://github.com/dmlc/xgboost/tree/master/demo>`_ for more code examples.
|
||||
|
||||
******
|
||||
Python
|
||||
******
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
import xgboost as xgb
|
||||
# read in data
|
||||
dtrain = xgb.DMatrix('demo/data/agaricus.txt.train')
|
||||
dtest = xgb.DMatrix('demo/data/agaricus.txt.test')
|
||||
# specify parameters via map
|
||||
param = {'max_depth':2, 'eta':1, 'silent':1, 'objective':'binary:logistic' }
|
||||
num_round = 2
|
||||
bst = xgb.train(param, dtrain, num_round)
|
||||
# make prediction
|
||||
preds = bst.predict(dtest)
|
||||
|
||||
***
|
||||
R
|
||||
***
|
||||
|
||||
.. code-block:: R
|
||||
|
||||
# load data
|
||||
data(agaricus.train, package='xgboost')
|
||||
data(agaricus.test, package='xgboost')
|
||||
train <- agaricus.train
|
||||
test <- agaricus.test
|
||||
# fit model
|
||||
bst <- xgboost(data = train$data, label = train$label, max.depth = 2, eta = 1, nround = 2,
|
||||
nthread = 2, objective = "binary:logistic")
|
||||
# predict
|
||||
pred <- predict(bst, test$data)
|
||||
|
||||
*****
|
||||
Julia
|
||||
*****
|
||||
|
||||
.. code-block:: julia
|
||||
|
||||
using XGBoost
|
||||
# read data
|
||||
train_X, train_Y = readlibsvm("demo/data/agaricus.txt.train", (6513, 126))
|
||||
test_X, test_Y = readlibsvm("demo/data/agaricus.txt.test", (1611, 126))
|
||||
# fit model
|
||||
num_round = 2
|
||||
bst = xgboost(train_X, num_round, label=train_Y, eta=1, max_depth=2)
|
||||
# predict
|
||||
pred = predict(bst, test_X)
|
||||
|
||||
*****
|
||||
Scala
|
||||
*****
|
||||
|
||||
.. code-block:: scala
|
||||
|
||||
import ml.dmlc.xgboost4j.scala.DMatrix
|
||||
import ml.dmlc.xgboost4j.scala.XGBoost
|
||||
|
||||
object XGBoostScalaExample {
|
||||
def main(args: Array[String]) {
|
||||
// read trainining data, available at xgboost/demo/data
|
||||
val trainData =
|
||||
new DMatrix("/path/to/agaricus.txt.train")
|
||||
// define parameters
|
||||
val paramMap = List(
|
||||
"eta" -> 0.1,
|
||||
"max_depth" -> 2,
|
||||
"objective" -> "binary:logistic").toMap
|
||||
// number of iterations
|
||||
val round = 2
|
||||
// train the model
|
||||
val model = XGBoost.train(trainData, paramMap, round)
|
||||
// run prediction
|
||||
val predTrain = model.predict(trainData)
|
||||
// save model to the file.
|
||||
model.saveModel("/local/path/to/model")
|
||||
}
|
||||
}
|
||||
@ -1,80 +0,0 @@
|
||||
# Get Started with XGBoost
|
||||
|
||||
This is a quick start tutorial showing snippets for you to quickly try out xgboost
|
||||
on the demo dataset on a binary classification task.
|
||||
|
||||
## Links to Helpful Other Resources
|
||||
- See [Installation Guide](../build.md) on how to install xgboost.
|
||||
- See [Text Input Format](../input_format.md) on using text format for specifying training/testing data.
|
||||
- See [How to pages](../how_to/index.md) on various tips on using xgboost.
|
||||
- See [Tutorials](../tutorials/index.md) on tutorials on specific tasks.
|
||||
- See [Learning to use XGBoost by Examples](../../demo) for more code examples.
|
||||
|
||||
## Python
|
||||
```python
|
||||
import xgboost as xgb
|
||||
# read in data
|
||||
dtrain = xgb.DMatrix('demo/data/agaricus.txt.train')
|
||||
dtest = xgb.DMatrix('demo/data/agaricus.txt.test')
|
||||
# specify parameters via map
|
||||
param = {'max_depth':2, 'eta':1, 'silent':1, 'objective':'binary:logistic' }
|
||||
num_round = 2
|
||||
bst = xgb.train(param, dtrain, num_round)
|
||||
# make prediction
|
||||
preds = bst.predict(dtest)
|
||||
```
|
||||
|
||||
## R
|
||||
|
||||
```r
|
||||
# load data
|
||||
data(agaricus.train, package='xgboost')
|
||||
data(agaricus.test, package='xgboost')
|
||||
train <- agaricus.train
|
||||
test <- agaricus.test
|
||||
# fit model
|
||||
bst <- xgboost(data = train$data, label = train$label, max.depth = 2, eta = 1, nround = 2,
|
||||
nthread = 2, objective = "binary:logistic")
|
||||
# predict
|
||||
pred <- predict(bst, test$data)
|
||||
```
|
||||
|
||||
## Julia
|
||||
```julia
|
||||
using XGBoost
|
||||
# read data
|
||||
train_X, train_Y = readlibsvm("demo/data/agaricus.txt.train", (6513, 126))
|
||||
test_X, test_Y = readlibsvm("demo/data/agaricus.txt.test", (1611, 126))
|
||||
# fit model
|
||||
num_round = 2
|
||||
bst = xgboost(train_X, num_round, label=train_Y, eta=1, max_depth=2)
|
||||
# predict
|
||||
pred = predict(bst, test_X)
|
||||
```
|
||||
|
||||
## Scala
|
||||
```scala
|
||||
import ml.dmlc.xgboost4j.scala.DMatrix
|
||||
import ml.dmlc.xgboost4j.scala.XGBoost
|
||||
|
||||
object XGBoostScalaExample {
|
||||
def main(args: Array[String]) {
|
||||
// read trainining data, available at xgboost/demo/data
|
||||
val trainData =
|
||||
new DMatrix("/path/to/agaricus.txt.train")
|
||||
// define parameters
|
||||
val paramMap = List(
|
||||
"eta" -> 0.1,
|
||||
"max_depth" -> 2,
|
||||
"objective" -> "binary:logistic").toMap
|
||||
// number of iterations
|
||||
val round = 2
|
||||
// train the model
|
||||
val model = XGBoost.train(trainData, paramMap, round)
|
||||
// run prediction
|
||||
val predTrain = model.predict(trainData)
|
||||
// save model to the file.
|
||||
model.saveModel("/local/path/to/model")
|
||||
}
|
||||
}
|
||||
```
|
||||
105
doc/gpu/index.md
105
doc/gpu/index.md
@ -1,105 +0,0 @@
|
||||
XGBoost GPU Support
|
||||
===================
|
||||
|
||||
This page contains information about GPU algorithms supported in XGBoost.
|
||||
To install GPU support, checkout the [build and installation instructions](../build.md).
|
||||
|
||||
# CUDA Accelerated Tree Construction Algorithms
|
||||
This plugin adds GPU accelerated tree construction and prediction algorithms to XGBoost.
|
||||
## Usage
|
||||
Specify the 'tree_method' parameter as one of the following algorithms.
|
||||
|
||||
### Algorithms
|
||||
|
||||
```eval_rst
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
| tree_method | Description |
|
||||
+==============+=======================================================================================================================================================================+
|
||||
| gpu_exact | The standard XGBoost tree construction algorithm. Performs exact search for splits. Slower and uses considerably more memory than 'gpu_hist' |
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
| gpu_hist | Equivalent to the XGBoost fast histogram algorithm. Much faster and uses considerably less memory. NOTE: Will run very slowly on GPUs older than Pascal architecture. |
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
```
|
||||
|
||||
### Supported parameters
|
||||
|
||||
```eval_rst
|
||||
.. |tick| unicode:: U+2714
|
||||
.. |cross| unicode:: U+2718
|
||||
|
||||
+----------------------+------------+-----------+
|
||||
| parameter | gpu_exact | gpu_hist |
|
||||
+======================+============+===========+
|
||||
| subsample | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| colsample_bytree | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| colsample_bylevel | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| max_bin | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| gpu_id | |tick| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| n_gpus | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| predictor | |tick| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| grow_policy | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
| monotone_constraints | |cross| | |tick| |
|
||||
+----------------------+------------+-----------+
|
||||
```
|
||||
|
||||
GPU accelerated prediction is enabled by default for the above mentioned 'tree_method' parameters but can be switched to CPU prediction by setting 'predictor':'cpu_predictor'. This could be useful if you want to conserve GPU memory. Likewise when using CPU algorithms, GPU accelerated prediction can be enabled by setting 'predictor':'gpu_predictor'.
|
||||
|
||||
The device ordinal can be selected using the 'gpu_id' parameter, which defaults to 0.
|
||||
|
||||
Multiple GPUs can be used with the grow_gpu_hist parameter using the n_gpus parameter. which defaults to 1. If this is set to -1 all available GPUs will be used. If gpu_id is specified as non-zero, the gpu device order is mod(gpu_id + i) % n_visible_devices for i=0 to n_gpus-1. As with GPU vs. CPU, multi-GPU will not always be faster than a single GPU due to PCI bus bandwidth that can limit performance.
|
||||
|
||||
This plugin currently works with the CLI, python and R - see installation guide for details.
|
||||
|
||||
Python example:
|
||||
```python
|
||||
param['gpu_id'] = 0
|
||||
param['max_bin'] = 16
|
||||
param['tree_method'] = 'gpu_hist'
|
||||
```
|
||||
## Benchmarks
|
||||
To run benchmarks on synthetic data for binary classification:
|
||||
```bash
|
||||
$ python tests/benchmark/benchmark.py
|
||||
```
|
||||
|
||||
Training time time on 1,000,000 rows x 50 columns with 500 boosting iterations and 0.25/0.75 test/train split on i7-6700K CPU @ 4.00GHz and Pascal Titan X.
|
||||
|
||||
```eval_rst
|
||||
+--------------+----------+
|
||||
| tree_method | Time (s) |
|
||||
+==============+==========+
|
||||
| gpu_hist | 13.87 |
|
||||
+--------------+----------+
|
||||
| hist | 63.55 |
|
||||
+--------------+----------+
|
||||
| gpu_exact | 161.08 |
|
||||
+--------------+----------+
|
||||
| exact | 1082.20 |
|
||||
+--------------+----------+
|
||||
|
||||
```
|
||||
|
||||
[See here](http://dmlc.ml/2016/12/14/GPU-accelerated-xgboost.html) for additional performance benchmarks of the 'gpu_exact' tree_method.
|
||||
|
||||
## References
|
||||
[Mitchell R, Frank E. (2017) Accelerating the XGBoost algorithm using GPU computing. PeerJ Computer Science 3:e127 https://doi.org/10.7717/peerj-cs.127](https://peerj.com/articles/cs-127/)
|
||||
|
||||
[Nvidia Parallel Forall: Gradient Boosting, Decision Trees and XGBoost with CUDA](https://devblogs.nvidia.com/parallelforall/gradient-boosting-decision-trees-xgboost-cuda/)
|
||||
|
||||
## Author
|
||||
Rory Mitchell
|
||||
Jonathan C. McKinney
|
||||
Shankara Rao Thejaswi Nanditale
|
||||
Vinay Deshpande
|
||||
... and the rest of the H2O.ai and NVIDIA team.
|
||||
|
||||
Please report bugs to the xgboost/issues page.
|
||||
|
||||
111
doc/gpu/index.rst
Normal file
111
doc/gpu/index.rst
Normal file
@ -0,0 +1,111 @@
|
||||
###################
|
||||
XGBoost GPU Support
|
||||
###################
|
||||
|
||||
This page contains information about GPU algorithms supported in XGBoost.
|
||||
To install GPU support, checkout the :doc:`/build`.
|
||||
|
||||
*********************************************
|
||||
CUDA Accelerated Tree Construction Algorithms
|
||||
*********************************************
|
||||
This plugin adds GPU accelerated tree construction and prediction algorithms to XGBoost.
|
||||
|
||||
Usage
|
||||
=====
|
||||
Specify the ``tree_method`` parameter as one of the following algorithms.
|
||||
|
||||
Algorithms
|
||||
----------
|
||||
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
| tree_method | Description |
|
||||
+==============+=======================================================================================================================================================================+
|
||||
| gpu_exact | The standard XGBoost tree construction algorithm. Performs exact search for splits. Slower and uses considerably more memory than ``gpu_hist``. |
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
| gpu_hist | Equivalent to the XGBoost fast histogram algorithm. Much faster and uses considerably less memory. NOTE: Will run very slowly on GPUs older than Pascal architecture. |
|
||||
+--------------+-----------------------------------------------------------------------------------------------------------------------------------------------------------------------+
|
||||
|
||||
Supported parameters
|
||||
--------------------
|
||||
|
||||
.. |tick| unicode:: U+2714
|
||||
.. |cross| unicode:: U+2718
|
||||
|
||||
+--------------------------+---------------+--------------+
|
||||
| parameter | ``gpu_exact`` | ``gpu_hist`` |
|
||||
+==========================+===============+==============+
|
||||
| ``subsample`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``colsample_bytree`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``colsample_bylevel`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``max_bin`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``gpu_id`` | |tick| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``n_gpus`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``predictor`` | |tick| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``grow_policy`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
| ``monotone_constraints`` | |cross| | |tick| |
|
||||
+--------------------------+---------------+--------------+
|
||||
|
||||
GPU accelerated prediction is enabled by default for the above mentioned ``tree_method`` parameters but can be switched to CPU prediction by setting ``predictor`` to ``cpu_predictor``. This could be useful if you want to conserve GPU memory. Likewise when using CPU algorithms, GPU accelerated prediction can be enabled by setting ``predictor`` to ``gpu_predictor``.
|
||||
|
||||
The device ordinal can be selected using the ``gpu_id`` parameter, which defaults to 0.
|
||||
|
||||
Multiple GPUs can be used with the ``gpu_hist`` tree method using the ``n_gpus`` parameter. which defaults to 1. If this is set to -1 all available GPUs will be used. If ``gpu_id`` is specified as non-zero, the gpu device order is ``mod(gpu_id + i) % n_visible_devices`` for ``i=0`` to ``n_gpus-1``. As with GPU vs. CPU, multi-GPU will not always be faster than a single GPU due to PCI bus bandwidth that can limit performance.
|
||||
|
||||
This plugin currently works with the CLI, python and R - see :doc:`/build` for details.
|
||||
|
||||
.. code-block:: python
|
||||
:caption: Python example
|
||||
|
||||
param['gpu_id'] = 0
|
||||
param['max_bin'] = 16
|
||||
param['tree_method'] = 'gpu_hist'
|
||||
|
||||
Benchmarks
|
||||
==========
|
||||
You can run benchmarks on synthetic data for binary classification:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
python tests/benchmark/benchmark.py
|
||||
|
||||
Training time time on 1,000,000 rows x 50 columns with 500 boosting iterations and 0.25/0.75 test/train split on i7-6700K CPU @ 4.00GHz and Pascal Titan X yields the following results:
|
||||
|
||||
+--------------+----------+
|
||||
| tree_method | Time (s) |
|
||||
+==============+==========+
|
||||
| gpu_hist | 13.87 |
|
||||
+--------------+----------+
|
||||
| hist | 63.55 |
|
||||
+--------------+----------+
|
||||
| gpu_exact | 161.08 |
|
||||
+--------------+----------+
|
||||
| exact | 1082.20 |
|
||||
+--------------+----------+
|
||||
|
||||
See `GPU Accelerated XGBoost <https://xgboost.ai/2016/12/14/GPU-accelerated-xgboost.html>`_ and `Updates to the XGBoost GPU algorithms <https://xgboost.ai/2018/07/04/gpu-xgboost-update.html>`_ for additional performance benchmarks of the ``gpu_exact`` and ``gpu_hist`` tree methods.
|
||||
|
||||
**********
|
||||
References
|
||||
**********
|
||||
`Mitchell R, Frank E. (2017) Accelerating the XGBoost algorithm using GPU computing. PeerJ Computer Science 3:e127 https://doi.org/10.7717/peerj-cs.127 <https://peerj.com/articles/cs-127/>`_
|
||||
|
||||
`Nvidia Parallel Forall: Gradient Boosting, Decision Trees and XGBoost with CUDA <https://devblogs.nvidia.com/parallelforall/gradient-boosting-decision-trees-xgboost-cuda/>`_
|
||||
|
||||
Authors
|
||||
=======
|
||||
* Rory Mitchell
|
||||
* Jonathan C. McKinney
|
||||
* Shankara Rao Thejaswi Nanditale
|
||||
* Vinay Deshpande
|
||||
* ... and the rest of the H2O.ai and NVIDIA team.
|
||||
|
||||
Please report bugs to the user forum https://discuss.xgboost.ai/.
|
||||
|
||||
@ -1,164 +0,0 @@
|
||||
Contribute to XGBoost
|
||||
=====================
|
||||
XGBoost has been developed and used by a group of active community members.
|
||||
Everyone is more than welcome to contribute. It is a way to make the project better and more accessible to more users.
|
||||
|
||||
- Please add your name to [CONTRIBUTORS.md](../../CONTRIBUTORS.md) after your patch has been merged.
|
||||
- Please also update [NEWS.md](../../NEWS.md) to add note on your changes to the API or added a new document.
|
||||
|
||||
Guidelines
|
||||
----------
|
||||
* [Submit Pull Request](#submit-pull-request)
|
||||
* [Git Workflow Howtos](#git-workflow-howtos)
|
||||
- [How to resolve conflict with master](#how-to-resolve-conflict-with-master)
|
||||
- [How to combine multiple commits into one](#how-to-combine-multiple-commits-into-one)
|
||||
- [What is the consequence of force push](#what-is-the-consequence-of-force-push)
|
||||
* [Document](#document)
|
||||
* [Testcases](#testcases)
|
||||
* [Examples](#examples)
|
||||
* [Core Library](#core-library)
|
||||
* [Python Package](#python-package)
|
||||
* [R Package](#r-package)
|
||||
|
||||
Submit Pull Request
|
||||
-------------------
|
||||
* Before submit, please rebase your code on the most recent version of master, you can do it by
|
||||
```bash
|
||||
git remote add upstream https://github.com/dmlc/xgboost
|
||||
git fetch upstream
|
||||
git rebase upstream/master
|
||||
```
|
||||
* If you have multiple small commits,
|
||||
it might be good to merge them together(use git rebase then squash) into more meaningful groups.
|
||||
* Send the pull request!
|
||||
- Fix the problems reported by automatic checks
|
||||
- If you are contributing a new module, consider add a testcase in [tests](../tests)
|
||||
|
||||
Git Workflow Howtos
|
||||
-------------------
|
||||
### How to resolve conflict with master
|
||||
- First rebase to most recent master
|
||||
```bash
|
||||
# The first two steps can be skipped after you do it once.
|
||||
git remote add upstream https://github.com/dmlc/xgboost
|
||||
git fetch upstream
|
||||
git rebase upstream/master
|
||||
```
|
||||
- The git may show some conflicts it cannot merge, say ```conflicted.py```.
|
||||
- Manually modify the file to resolve the conflict.
|
||||
- After you resolved the conflict, mark it as resolved by
|
||||
```bash
|
||||
git add conflicted.py
|
||||
```
|
||||
- Then you can continue rebase by
|
||||
```bash
|
||||
git rebase --continue
|
||||
```
|
||||
- Finally push to your fork, you may need to force push here.
|
||||
```bash
|
||||
git push --force
|
||||
```
|
||||
|
||||
### How to combine multiple commits into one
|
||||
Sometimes we want to combine multiple commits, especially when later commits are only fixes to previous ones,
|
||||
to create a PR with set of meaningful commits. You can do it by following steps.
|
||||
- Before doing so, configure the default editor of git if you haven't done so before.
|
||||
```bash
|
||||
git config core.editor the-editor-you-like
|
||||
```
|
||||
- Assume we want to merge last 3 commits, type the following commands
|
||||
```bash
|
||||
git rebase -i HEAD~3
|
||||
```
|
||||
- It will pop up an text editor. Set the first commit as ```pick```, and change later ones to ```squash```.
|
||||
- After you saved the file, it will pop up another text editor to ask you modify the combined commit message.
|
||||
- Push the changes to your fork, you need to force push.
|
||||
```bash
|
||||
git push --force
|
||||
```
|
||||
|
||||
### What is the consequence of force push
|
||||
The previous two tips requires force push, this is because we altered the path of the commits.
|
||||
It is fine to force push to your own fork, as long as the commits changed are only yours.
|
||||
|
||||
Documents
|
||||
---------
|
||||
* The document is created using sphinx and [recommonmark](http://recommonmark.readthedocs.org/en/latest/)
|
||||
* You can build document locally to see the effect.
|
||||
|
||||
Testcases
|
||||
---------
|
||||
* All the testcases are in [tests](../tests)
|
||||
* We use python nose for python test cases.
|
||||
|
||||
Examples
|
||||
--------
|
||||
* Usecases and examples will be in [demo](../demo)
|
||||
* We are super excited to hear about your story, if you have blogposts,
|
||||
tutorials code solutions using xgboost, please tell us and we will add
|
||||
a link in the example pages.
|
||||
|
||||
Core Library
|
||||
------------
|
||||
- Follow Google C style for C++.
|
||||
- We use doxygen to document all the interface code.
|
||||
- You can reproduce the linter checks by typing ```make lint```
|
||||
|
||||
Python Package
|
||||
--------------
|
||||
- Always add docstring to the new functions in numpydoc format.
|
||||
- You can reproduce the linter checks by typing ```make lint```
|
||||
|
||||
R Package
|
||||
---------
|
||||
### Code Style
|
||||
- We follow Google's C++ Style guide on C++ code.
|
||||
- This is mainly to be consistent with the rest of the project.
|
||||
- Another reason is we will be able to check style automatically with a linter.
|
||||
- You can check the style of the code by typing the following command at root folder.
|
||||
```bash
|
||||
make rcpplint
|
||||
```
|
||||
- When needed, you can disable the linter warning of certain line with ```// NOLINT(*)``` comments.
|
||||
- We use [roxygen](https://cran.r-project.org/web/packages/roxygen2/vignettes/roxygen2.html) for documenting the R package.
|
||||
|
||||
### Rmarkdown Vignettes
|
||||
Rmarkdown vignettes are placed in [R-package/vignettes](../R-package/vignettes)
|
||||
These Rmarkdown files are not compiled. We host the compiled version on [doc/R-package](R-package)
|
||||
|
||||
The following steps are followed to add a new Rmarkdown vignettes:
|
||||
- Add the original rmarkdown to ```R-package/vignettes```
|
||||
- Modify ```doc/R-package/Makefile``` to add the markdown files to be build
|
||||
- Clone the [dmlc/web-data](https://github.com/dmlc/web-data) repo to folder ```doc```
|
||||
- Now type the following command on ```doc/R-package```
|
||||
```bash
|
||||
make the-markdown-to-make.md
|
||||
```
|
||||
- This will generate the markdown, as well as the figures into ```doc/web-data/xgboost/knitr```
|
||||
- Modify the ```doc/R-package/index.md``` to point to the generated markdown.
|
||||
- Add the generated figure to the ```dmlc/web-data``` repo.
|
||||
- If you already cloned the repo to doc, this means a ```git add```
|
||||
- Create PR for both the markdown and ```dmlc/web-data```
|
||||
- You can also build the document locally by typing the following command at ```doc```
|
||||
```bash
|
||||
make html
|
||||
```
|
||||
The reason we do this is to avoid exploded repo size due to generated images sizes.
|
||||
|
||||
### R package versioning
|
||||
Since version 0.6.4.3, we have adopted a versioning system that uses an ```x.y.z``` (or ```core_major.core_minor.cran_release```)
|
||||
format for CRAN releases and an ```x.y.z.p``` (or ```core_major.core_minor.cran_release.patch```) format for development patch versions.
|
||||
This approach is similar to the one described in Yihui Xie's
|
||||
[blog post on R Package Versioning](https://yihui.name/en/2013/06/r-package-versioning/),
|
||||
except we need an additional field to accomodate the ```x.y``` core library version.
|
||||
|
||||
Each new CRAN release bumps up the 3rd field, while developments in-between CRAN releases
|
||||
would be marked by an additional 4th field on the top of an existing CRAN release version.
|
||||
Some additional consideration is needed when the core library version changes.
|
||||
E.g., after the core changes from 0.6 to 0.7, the R package development version would become 0.7.0.1, working towards
|
||||
a 0.7.1 CRAN release. The 0.7.0 would not be released to CRAN, unless it would require almost no additional development.
|
||||
|
||||
### Registering native routines in R
|
||||
According to [R extension manual](https://cran.r-project.org/doc/manuals/r-release/R-exts.html#Registering-native-routines),
|
||||
it is good practice to register native routines and to disable symbol search. When any changes or additions are made to the
|
||||
C++ interface of the R package, please make corresponding changes in ```src/init.c``` as well.
|
||||
@ -1,42 +0,0 @@
|
||||
Using XGBoost External Memory Version(beta)
|
||||
===========================================
|
||||
There is no big difference between using external memory version and in-memory version.
|
||||
The only difference is the filename format.
|
||||
|
||||
The external memory version takes in the following filename format
|
||||
```
|
||||
filename#cacheprefix
|
||||
```
|
||||
|
||||
The ```filename``` is the normal path to libsvm file you want to load in, ```cacheprefix``` is a
|
||||
path to a cache file that xgboost will use for external memory cache.
|
||||
|
||||
The following code was extracted from [../../demo/guide-python/external_memory.py](../../demo/guide-python/external_memory.py)
|
||||
```python
|
||||
dtrain = xgb.DMatrix('../data/agaricus.txt.train#dtrain.cache')
|
||||
```
|
||||
You can find that there is additional ```#dtrain.cache``` following the libsvm file, this is the name of cache file.
|
||||
For CLI version, simply use ```"../data/agaricus.txt.train#dtrain.cache"``` in filename.
|
||||
|
||||
Performance Note
|
||||
----------------
|
||||
* the parameter ```nthread``` should be set to number of ***real*** cores
|
||||
- Most modern CPU offer hyperthreading, which means you can have a 4 core cpu with 8 threads
|
||||
- Set nthread to be 4 for maximum performance in such case
|
||||
|
||||
Distributed Version
|
||||
-------------------
|
||||
The external memory mode naturally works on distributed version, you can simply set path like
|
||||
```
|
||||
data = "hdfs://path-to-data/#dtrain.cache"
|
||||
```
|
||||
xgboost will cache the data to the local position. When you run on YARN, the current folder is temporal
|
||||
so that you can directly use ```dtrain.cache``` to cache to current folder.
|
||||
|
||||
|
||||
Usage Note
|
||||
----------
|
||||
* This is a experimental version
|
||||
- If you like to try and test it, report results to https://github.com/dmlc/xgboost/issues/244
|
||||
* Currently only importing from libsvm format is supported
|
||||
- Contribution of ingestion from other common external memory data source is welcomed
|
||||
@ -1,18 +0,0 @@
|
||||
# XGBoost How To
|
||||
|
||||
This page contains guidelines to use and develop XGBoost.
|
||||
|
||||
## Installation
|
||||
- [How to Install XGBoost](../build.md)
|
||||
|
||||
## Use XGBoost in Specific Ways
|
||||
- [Text input format](../input_format.md)
|
||||
- [Parameter tuning guide](param_tuning.md)
|
||||
- [Use out of core computation for large dataset](external_memory.md)
|
||||
- [Use XGBoost GPU algorithms](../gpu/index.md)
|
||||
|
||||
## Develop and Hack XGBoost
|
||||
- [Contribute to XGBoost](contribute.md)
|
||||
|
||||
## Frequently Ask Questions
|
||||
- [FAQ](../faq.md)
|
||||
16
doc/index.md
16
doc/index.md
@ -1,16 +0,0 @@
|
||||
XGBoost Documentation
|
||||
=====================
|
||||
This document is hosted at http://xgboost.readthedocs.org/. You can also browse most of the documents in github directly.
|
||||
|
||||
|
||||
These are used to generate the index used in search.
|
||||
|
||||
* [Python Package Document](python/index.md)
|
||||
* [R Package Document](R-package/index.md)
|
||||
* [Java/Scala Package Document](jvm/index.md)
|
||||
* [Julia Package Document](julia/index.md)
|
||||
* [CLI Package Document](cli/index.md)
|
||||
* [GPU Support Document](gpu/index.md)
|
||||
- [Howto Documents](how_to/index.md)
|
||||
- [Get Started Documents](get_started/index.md)
|
||||
- [Tutorials](tutorials/index.md)
|
||||
30
doc/index.rst
Normal file
30
doc/index.rst
Normal file
@ -0,0 +1,30 @@
|
||||
#####################
|
||||
XGBoost Documentation
|
||||
#####################
|
||||
|
||||
**XGBoost** is an optimized distributed gradient boosting library designed to be highly **efficient**, **flexible** and **portable**.
|
||||
It implements machine learning algorithms under the `Gradient Boosting <https://en.wikipedia.org/wiki/Gradient_boosting>`_ framework.
|
||||
XGBoost provides a parallel tree boosting (also known as GBDT, GBM) that solve many data science problems in a fast and accurate way.
|
||||
The same code runs on major distributed environment (Hadoop, SGE, MPI) and can solve problems beyond billions of examples.
|
||||
|
||||
********
|
||||
Contents
|
||||
********
|
||||
|
||||
.. toctree::
|
||||
:maxdepth: 2
|
||||
:titlesonly:
|
||||
|
||||
build
|
||||
get_started
|
||||
tutorials/index
|
||||
faq
|
||||
XGBoost User Forum <https://discuss.xgboost.ai>
|
||||
GPU support <gpu/index>
|
||||
parameter
|
||||
Python package <python/index>
|
||||
R package <R-package/index>
|
||||
JVM package <jvm/index>
|
||||
Julia package <julia>
|
||||
CLI interface <cli>
|
||||
contribute
|
||||
@ -1,93 +0,0 @@
|
||||
Text Input Format of DMatrix
|
||||
============================
|
||||
|
||||
## Basic Input Format
|
||||
XGBoost currently supports two text formats for ingesting data: LibSVM and CSV. The rest of this document will describe the LibSVM format. (See [here](https://en.wikipedia.org/wiki/Comma-separated_values) for a description of the CSV format.)
|
||||
|
||||
For training or predicting, XGBoost takes an instance file with the format as below:
|
||||
|
||||
train.txt
|
||||
```
|
||||
1 101:1.2 102:0.03
|
||||
0 1:2.1 10001:300 10002:400
|
||||
0 0:1.3 1:0.3
|
||||
1 0:0.01 1:0.3
|
||||
0 0:0.2 1:0.3
|
||||
```
|
||||
Each line represent a single instance, and in the first line '1' is the instance label,'101' and '102' are feature indices, '1.2' and '0.03' are feature values. In the binary classification case, '1' is used to indicate positive samples, and '0' is used to indicate negative samples. We also support probability values in [0,1] as label, to indicate the probability of the instance being positive.
|
||||
|
||||
Auxiliary Files for Additional Information
|
||||
------------------------------------------
|
||||
**Note: all information below is applicable only to single-node version of the package.** If you'd like to perform distributed training with multiple nodes, skip to the next section.
|
||||
|
||||
### Group Input Format
|
||||
For [ranking task](../demo/rank), XGBoost supports the group input format. In ranking task, instances are categorized into *query groups* in real world scenarios. For example, in the learning to rank web pages scenario, the web page instances are grouped by their queries. XGBoost requires an file that indicates the group information. For example, if the instance file is the "train.txt" shown above, the group file should be named "train.txt.group" and be of the following format:
|
||||
|
||||
train.txt.group
|
||||
```
|
||||
2
|
||||
3
|
||||
```
|
||||
This means that, the data set contains 5 instances, and the first two instances are in a group and the other three are in another group. The numbers in the group file are actually indicating the number of instances in each group in the instance file in order.
|
||||
At the time of configuration, you do not have to indicate the path of the group file. If the instance file name is "xxx", XGBoost will check whether there is a file named "xxx.group" in the same directory.
|
||||
|
||||
### Instance Weight File
|
||||
Instances in the training data may be assigned weights to differentiate relative importance among them. For example, if we provide an instance weight file for the "train.txt" file in the example as below:
|
||||
|
||||
train.txt.weight
|
||||
```
|
||||
1
|
||||
0.5
|
||||
0.5
|
||||
1
|
||||
0.5
|
||||
```
|
||||
It means that XGBoost will emphasize more on the first and fourth instance (i.e. the positive instances) while training.
|
||||
The configuration is similar to configuring the group information. If the instance file name is "xxx", XGBoost will look for a file named "xxx.weight" in the same directory. If the file exists, the instance weights will be extracted and used at the time of training.
|
||||
|
||||
NOTE. If you choose to save the training data as a binary buffer (using [save_binary()](http://xgboost.readthedocs.io/en/latest/python/python_api.html#xgboost.DMatrix.save_binary)), keep in mind that the resulting binary buffer file will include the instance weights. To update the weights, use [the set_weight() function](http://xgboost.readthedocs.io/en/latest/python/python_api.html#xgboost.DMatrix.set_weight).
|
||||
|
||||
### Initial Margin File
|
||||
XGBoost supports providing each instance an initial margin prediction. For example, if we have a initial prediction using logistic regression for "train.txt" file, we can create the following file:
|
||||
|
||||
train.txt.base_margin
|
||||
```
|
||||
-0.4
|
||||
1.0
|
||||
3.4
|
||||
```
|
||||
XGBoost will take these values as initial margin prediction and boost from that. An important note about base_margin is that it should be margin prediction before transformation, so if you are doing logistic loss, you will need to put in value before logistic transformation. If you are using XGBoost predictor, use pred_margin=1 to output margin values.
|
||||
|
||||
Embedding additional information inside LibSVM file
|
||||
---------------------------------------------------
|
||||
**This section is applicable to both single- and multiple-node settings.**
|
||||
|
||||
### Query ID Columns
|
||||
This is most useful for [ranking task](../demo/rank), where the instances are grouped into query groups. You may embed query group ID for each instance in the LibSVM file by adding a token of form `qid:xx` in each row:
|
||||
|
||||
train.txt
|
||||
```
|
||||
1 qid:1 101:1.2 102:0.03
|
||||
0 qid:1 1:2.1 10001:300 10002:400
|
||||
0 qid:2 0:1.3 1:0.3
|
||||
1 qid:2 0:0.01 1:0.3
|
||||
0 qid:3 0:0.2 1:0.3
|
||||
1 qid:3 3:-0.1 10:-0.3
|
||||
0 qid:3 6:0.2 10:0.15
|
||||
```
|
||||
Keep in mind the following restrictions:
|
||||
* It is not allowed to specify query ID's for some instances but not for others. Either every row is assigned query ID's or none at all.
|
||||
* The rows have to be sorted in ascending order by the query IDs. So, for instance, you may not have one row having large query ID than any of the following rows.
|
||||
|
||||
### Instance weights
|
||||
You may specify instance weights in the LibSVM file by appending each instance label with the corresponding weight in the form of `[label]:[weight]`, as shown by the following example:
|
||||
|
||||
train.txt
|
||||
```
|
||||
1:1.0 101:1.2 102:0.03
|
||||
0:0.5 1:2.1 10001:300 10002:400
|
||||
0:0.5 0:1.3 1:0.3
|
||||
1:1.0 0:0.01 1:0.3
|
||||
0:0.5 0:0.2 1:0.3
|
||||
```
|
||||
where the negative instances are assigned half weights compared to the positive instances.
|
||||
5
doc/julia.rst
Normal file
5
doc/julia.rst
Normal file
@ -0,0 +1,5 @@
|
||||
##########
|
||||
XGBoost.jl
|
||||
##########
|
||||
|
||||
See `XGBoost.jl Project page <https://github.com/dmlc/XGBoost.jl>`_.
|
||||
@ -1,3 +0,0 @@
|
||||
# XGBoost.jl
|
||||
|
||||
See [XGBoost.jl Project page](https://github.com/dmlc/XGBoost.jl)
|
||||
134
doc/jvm/index.md
134
doc/jvm/index.md
@ -1,134 +0,0 @@
|
||||
XGBoost JVM Package
|
||||
===================
|
||||
[](https://travis-ci.org/dmlc/xgboost)
|
||||
[](../LICENSE)
|
||||
|
||||
You have found the XGBoost JVM Package!
|
||||
|
||||
Installation
|
||||
------------
|
||||
|
||||
#### Installation from source
|
||||
|
||||
Building XGBoost4J using Maven requires Maven 3 or newer, Java 7+ and CMake 3.2+ for compiling the JNI bindings.
|
||||
|
||||
Before you install XGBoost4J, you need to define environment variable `JAVA_HOME` as your JDK directory to ensure that your compiler can find `jni.h` correctly, since XGBoost4J relies on JNI to implement the interaction between the JVM and native libraries.
|
||||
|
||||
After your `JAVA_HOME` is defined correctly, it is as simple as run `mvn package` under jvm-packages directory to install XGBoost4J. You can also skip the tests by running `mvn -DskipTests=true package`, if you are sure about the correctness of your local setup.
|
||||
|
||||
To publish the artifacts to your local maven repository, run
|
||||
|
||||
mvn install
|
||||
|
||||
Or, if you would like to skip tests, run
|
||||
|
||||
mvn -DskipTests install
|
||||
|
||||
This command will publish the xgboost binaries, the compiled java classes as well as the java sources to your local repository. Then you can use XGBoost4J in your Java projects by including the following dependency in `pom.xml`:
|
||||
|
||||
<b>maven</b>
|
||||
|
||||
```
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_source_version_num</version>
|
||||
</dependency>
|
||||
```
|
||||
|
||||
For sbt, please add the repository and dependency in build.sbt as following:
|
||||
|
||||
<b>sbt</b>
|
||||
```sbt
|
||||
resolvers += "Local Maven Repository" at "file://"+Path.userHome.absolutePath+"/.m2/repository"
|
||||
|
||||
"ml.dmlc" % "xgboost4j" % "latest_source_version_num"
|
||||
```
|
||||
|
||||
|
||||
#### Installation from maven repo
|
||||
|
||||
### Access release version
|
||||
|
||||
<b>maven</b>
|
||||
|
||||
```
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_version_num</version>
|
||||
</dependency>
|
||||
```
|
||||
|
||||
<b>sbt</b>
|
||||
```sbt
|
||||
"ml.dmlc" % "xgboost4j" % "latest_version_num"
|
||||
```
|
||||
|
||||
For the latest release version number, please check [here](https://github.com/dmlc/xgboost/releases).
|
||||
|
||||
if you want to use `xgboost4j-spark`, you just need to replace xgboost4j with `xgboost4j-spark`
|
||||
|
||||
### Access SNAPSHOT version
|
||||
|
||||
You need to add github as repo:
|
||||
|
||||
<b>maven</b>:
|
||||
|
||||
```xml
|
||||
<repository>
|
||||
<id>GitHub Repo</id>
|
||||
<name>GitHub Repo</name>
|
||||
<url>https://raw.githubusercontent.com/CodingCat/xgboost/maven-repo/</url>
|
||||
</repository>
|
||||
```
|
||||
|
||||
<b>sbt</b>:
|
||||
|
||||
```sbt
|
||||
resolvers += "GitHub Repo" at "https://raw.githubusercontent.com/CodingCat/xgboost/maven-repo/"
|
||||
```
|
||||
|
||||
the add dependency as following:
|
||||
|
||||
<b>maven</b>
|
||||
|
||||
```
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_version_num</version>
|
||||
</dependency>
|
||||
```
|
||||
|
||||
<b>sbt</b>
|
||||
```sbt
|
||||
"ml.dmlc" % "xgboost4j" % "latest_version_num"
|
||||
```
|
||||
|
||||
For the latest release version number, please check [here](https://github.com/CodingCat/xgboost/tree/maven-repo/ml/dmlc/xgboost4j).
|
||||
|
||||
if you want to use `xgboost4j-spark`, you just need to replace xgboost4j with `xgboost4j-spark`
|
||||
|
||||
After integrating with Dataframe/Dataset APIs of Spark 2.0, XGBoost4J-Spark only supports compile with Spark 2.x. You can build XGBoost4J-Spark as a component of XGBoost4J by running `mvn package`, and you can specify the version of spark with `mvn -Dspark.version=2.0.0 package`. (To continue working with Spark 1.x, the users are supposed to update pom.xml by modifying the properties like `spark.version`, `scala.version`, and `scala.binary.version`. Users also need to change the implementation by replacing SparkSession with SQLContext and the type of API parameters from Dataset[_] to Dataframe)
|
||||
|
||||
#### Enabling OpenMP for Mac OS
|
||||
If you are on Mac OS and using a compiler that supports OpenMP, you need to go to the file `xgboost/jvm-packages/create_jni.py` and comment out the line
|
||||
```python
|
||||
CONFIG["USE_OPENMP"] = "OFF"
|
||||
```
|
||||
in order to get the benefit of multi-threading.
|
||||
|
||||
Contents
|
||||
--------
|
||||
* [Java Overview Tutorial](java_intro.md)
|
||||
|
||||
Resources
|
||||
---------
|
||||
* [Code Examples](https://github.com/dmlc/xgboost/tree/master/jvm-packages/xgboost4j-example)
|
||||
* [Java API Docs](http://dmlc.ml/docs/javadocs/index.html)
|
||||
|
||||
## Scala API Docs
|
||||
* [XGBoost4J](http://dmlc.ml/docs/scaladocs/xgboost4j/index.html)
|
||||
* [XGBoost4J-Spark](http://dmlc.ml/docs/scaladocs/xgboost4j-spark/index.html)
|
||||
* [XGBoost4J-Flink](http://dmlc.ml/docs/scaladocs/xgboost4j-flink/index.html)
|
||||
145
doc/jvm/index.rst
Normal file
145
doc/jvm/index.rst
Normal file
@ -0,0 +1,145 @@
|
||||
###################
|
||||
XGBoost JVM Package
|
||||
###################
|
||||
|
||||
.. raw:: html
|
||||
|
||||
<a href="https://travis-ci.org/dmlc/xgboost">
|
||||
<img alt="Build Status" src="https://travis-ci.org/dmlc/xgboost.svg?branch=master">
|
||||
</a>
|
||||
<a href="https://github.com/dmlc/xgboost/blob/master/LICENSE">
|
||||
<img alt="GitHub license" src="http://dmlc.github.io/img/apache2.svg">
|
||||
</a>
|
||||
|
||||
You have found the XGBoost JVM Package!
|
||||
|
||||
************
|
||||
Installation
|
||||
************
|
||||
|
||||
Installation from source
|
||||
========================
|
||||
|
||||
Building XGBoost4J using Maven requires Maven 3 or newer, Java 7+ and CMake 3.2+ for compiling the JNI bindings.
|
||||
|
||||
Before you install XGBoost4J, you need to define environment variable ``JAVA_HOME`` as your JDK directory to ensure that your compiler can find ``jni.h`` correctly, since XGBoost4J relies on JNI to implement the interaction between the JVM and native libraries.
|
||||
|
||||
After your ``JAVA_HOME`` is defined correctly, it is as simple as run ``mvn package`` under jvm-packages directory to install XGBoost4J. You can also skip the tests by running ``mvn -DskipTests=true package``, if you are sure about the correctness of your local setup.
|
||||
|
||||
To publish the artifacts to your local maven repository, run
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mvn install
|
||||
|
||||
Or, if you would like to skip tests, run
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
mvn -DskipTests install
|
||||
|
||||
This command will publish the xgboost binaries, the compiled java classes as well as the java sources to your local repository. Then you can use XGBoost4J in your Java projects by including the following dependency in ``pom.xml``:
|
||||
|
||||
.. code-block:: xml
|
||||
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_source_version_num</version>
|
||||
</dependency>
|
||||
|
||||
For sbt, please add the repository and dependency in build.sbt as following:
|
||||
|
||||
.. code-block:: scala
|
||||
|
||||
resolvers += "Local Maven Repository" at "file://"+Path.userHome.absolutePath+"/.m2/repository"
|
||||
|
||||
"ml.dmlc" % "xgboost4j" % "latest_source_version_num"
|
||||
|
||||
Installation from maven repo
|
||||
============================
|
||||
|
||||
Access release version
|
||||
----------------------
|
||||
|
||||
.. code-block:: xml
|
||||
:caption: maven
|
||||
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_version_num</version>
|
||||
</dependency>
|
||||
|
||||
.. code-block:: scala
|
||||
:caption: sbt
|
||||
|
||||
"ml.dmlc" % "xgboost4j" % "latest_version_num"
|
||||
|
||||
For the latest release version number, please check `here <https://github.com/dmlc/xgboost/releases>`_.
|
||||
|
||||
if you want to use XGBoost4J-Spark, you just need to replace ``xgboost4j`` with ``xgboost4j-spark``.
|
||||
|
||||
Access SNAPSHOT version
|
||||
-----------------------
|
||||
|
||||
You need to add GitHub as repo:
|
||||
|
||||
.. code-block:: xml
|
||||
:caption: maven
|
||||
|
||||
<repository>
|
||||
<id>GitHub Repo</id>
|
||||
<name>GitHub Repo</name>
|
||||
<url>https://raw.githubusercontent.com/CodingCat/xgboost/maven-repo/</url>
|
||||
</repository>
|
||||
|
||||
.. code-block:: scala
|
||||
:caption: sbt
|
||||
|
||||
resolvers += "GitHub Repo" at "https://raw.githubusercontent.com/CodingCat/xgboost/maven-repo/"
|
||||
|
||||
Then add dependency as following:
|
||||
|
||||
.. code-block:: xml
|
||||
:caption: maven
|
||||
|
||||
<dependency>
|
||||
<groupId>ml.dmlc</groupId>
|
||||
<artifactId>xgboost4j</artifactId>
|
||||
<version>latest_version_num</version>
|
||||
</dependency>
|
||||
|
||||
.. code-block:: scala
|
||||
:caption: sbt
|
||||
|
||||
"ml.dmlc" % "xgboost4j" % "latest_version_num"
|
||||
|
||||
For the latest release version number, please check `here <https://github.com/CodingCat/xgboost/tree/maven-repo/ml/dmlc/xgboost4j>`_.
|
||||
|
||||
if you want to use XGBoost4J-Spark, you just need to replace ``xgboost4j`` with ``xgboost4j-spark``.
|
||||
|
||||
After integrating with Dataframe/Dataset APIs of Spark 2.0, XGBoost4J-Spark only supports compile with Spark 2.x. You can build XGBoost4J-Spark as a component of XGBoost4J by running ``mvn package``, and you can specify the version of spark with ``mvn -Dspark.version=2.0.0 package``. (To continue working with Spark 1.x, the users are supposed to update pom.xml by modifying the properties like ``spark.version``, ``scala.version``, and ``scala.binary.version``. Users also need to change the implementation by replacing ``SparkSession`` with ``SQLContext`` and the type of API parameters from ``Dataset[_]`` to ``Dataframe``)
|
||||
|
||||
Enabling OpenMP for Mac OS
|
||||
--------------------------
|
||||
If you are on Mac OS and using a compiler that supports OpenMP, you need to go to the file ``xgboost/jvm-packages/create_jni.py`` and comment out the line
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
CONFIG["USE_OPENMP"] = "OFF"
|
||||
|
||||
in order to get the benefit of multi-threading.
|
||||
|
||||
********
|
||||
Contents
|
||||
********
|
||||
|
||||
.. toctree::
|
||||
|
||||
Java Overview Tutorial <java_intro>
|
||||
Code Examples <https://github.com/dmlc/xgboost/tree/master/jvm-packages/xgboost4j-example>
|
||||
XGBoost4J Java API <http://dmlc.ml/docs/javadocs/index.html>
|
||||
XGBoost4J Scala API <http://dmlc.ml/docs/scaladocs/xgboost4j/index.html>
|
||||
XGBoost4J-Spark Scala API <http://dmlc.ml/docs/scaladocs/xgboost4j-spark/index.html>
|
||||
XGBoost4J-Flink Scala API <http://dmlc.ml/docs/scaladocs/xgboost4j-flink/index.html>
|
||||
@ -1,143 +0,0 @@
|
||||
XGBoost4J Java API
|
||||
==================
|
||||
This tutorial introduces
|
||||
|
||||
## Data Interface
|
||||
Like the xgboost python module, xgboost4j uses ```DMatrix``` to handle data,
|
||||
libsvm txt format file, sparse matrix in CSR/CSC format, and dense matrix is
|
||||
supported.
|
||||
|
||||
* To import ```DMatrix``` :
|
||||
```java
|
||||
import org.dmlc.xgboost4j.DMatrix;
|
||||
```
|
||||
|
||||
* To load libsvm text format file, the usage is like :
|
||||
```java
|
||||
DMatrix dmat = new DMatrix("train.svm.txt");
|
||||
```
|
||||
|
||||
* To load sparse matrix in CSR/CSC format is a little complicated, the usage is like :
|
||||
suppose a sparse matrix :
|
||||
1 0 2 0
|
||||
4 0 0 3
|
||||
3 1 2 0
|
||||
|
||||
for CSR format
|
||||
```java
|
||||
long[] rowHeaders = new long[] {0,2,4,7};
|
||||
float[] data = new float[] {1f,2f,4f,3f,3f,1f,2f};
|
||||
int[] colIndex = new int[] {0,2,0,3,0,1,2};
|
||||
DMatrix dmat = new DMatrix(rowHeaders, colIndex, data, DMatrix.SparseType.CSR);
|
||||
```
|
||||
|
||||
for CSC format
|
||||
```java
|
||||
long[] colHeaders = new long[] {0,3,4,6,7};
|
||||
float[] data = new float[] {1f,4f,3f,1f,2f,2f,3f};
|
||||
int[] rowIndex = new int[] {0,1,2,2,0,2,1};
|
||||
DMatrix dmat = new DMatrix(colHeaders, rowIndex, data, DMatrix.SparseType.CSC);
|
||||
```
|
||||
|
||||
* To load 3*2 dense matrix, the usage is like :
|
||||
suppose a matrix :
|
||||
1 2
|
||||
3 4
|
||||
5 6
|
||||
|
||||
```java
|
||||
float[] data = new float[] {1f,2f,3f,4f,5f,6f};
|
||||
int nrow = 3;
|
||||
int ncol = 2;
|
||||
float missing = 0.0f;
|
||||
DMatrix dmat = new Matrix(data, nrow, ncol, missing);
|
||||
```
|
||||
|
||||
* To set weight :
|
||||
```java
|
||||
float[] weights = new float[] {1f,2f,1f};
|
||||
dmat.setWeight(weights);
|
||||
```
|
||||
|
||||
## Setting Parameters
|
||||
* in xgboost4j any ```Iterable<Entry<String, Object>>``` object could be used as parameters.
|
||||
|
||||
* to set parameters, for non-multiple value params, you can simply use entrySet of an Map:
|
||||
```java
|
||||
Map<String, Object> paramMap = new HashMap<>() {
|
||||
{
|
||||
put("eta", 1.0);
|
||||
put("max_depth", 2);
|
||||
put("silent", 1);
|
||||
put("objective", "binary:logistic");
|
||||
put("eval_metric", "logloss");
|
||||
}
|
||||
};
|
||||
Iterable<Entry<String, Object>> params = paramMap.entrySet();
|
||||
```
|
||||
* for the situation that multiple values with same param key, List<Entry<String, Object>> would be a good choice, e.g. :
|
||||
```java
|
||||
List<Entry<String, Object>> params = new ArrayList<Entry<String, Object>>() {
|
||||
{
|
||||
add(new SimpleEntry<String, Object>("eta", 1.0));
|
||||
add(new SimpleEntry<String, Object>("max_depth", 2.0));
|
||||
add(new SimpleEntry<String, Object>("silent", 1));
|
||||
add(new SimpleEntry<String, Object>("objective", "binary:logistic"));
|
||||
}
|
||||
};
|
||||
```
|
||||
|
||||
## Training Model
|
||||
With parameters and data, you are able to train a booster model.
|
||||
* Import ```Trainer``` and ```Booster``` :
|
||||
```java
|
||||
import org.dmlc.xgboost4j.Booster;
|
||||
import org.dmlc.xgboost4j.util.Trainer;
|
||||
```
|
||||
|
||||
* Training
|
||||
```java
|
||||
DMatrix trainMat = new DMatrix("train.svm.txt");
|
||||
DMatrix validMat = new DMatrix("valid.svm.txt");
|
||||
//specify a watchList to see the performance
|
||||
//any Iterable<Entry<String, DMatrix>> object could be used as watchList
|
||||
List<Entry<String, DMatrix>> watchs = new ArrayList<>();
|
||||
watchs.add(new SimpleEntry<>("train", trainMat));
|
||||
watchs.add(new SimpleEntry<>("test", testMat));
|
||||
int round = 2;
|
||||
Booster booster = Trainer.train(params, trainMat, round, watchs, null, null);
|
||||
```
|
||||
|
||||
* Saving model
|
||||
After training, you can save model and dump it out.
|
||||
```java
|
||||
booster.saveModel("model.bin");
|
||||
```
|
||||
|
||||
* Dump Model and Feature Map
|
||||
```java
|
||||
booster.dumpModel("modelInfo.txt", false)
|
||||
//dump with featureMap
|
||||
booster.dumpModel("modelInfo.txt", "featureMap.txt", false)
|
||||
```
|
||||
|
||||
* Load a model
|
||||
```java
|
||||
Params param = new Params() {
|
||||
{
|
||||
put("silent", 1);
|
||||
put("nthread", 6);
|
||||
}
|
||||
};
|
||||
Booster booster = new Booster(param, "model.bin");
|
||||
```
|
||||
|
||||
## Prediction
|
||||
after training and loading a model, you use it to predict other data, the predict results will be a two-dimension float array (nsample, nclass), for predict leaf, it would be (nsample, nclass*ntrees)
|
||||
```java
|
||||
DMatrix dtest = new DMatrix("test.svm.txt");
|
||||
//predict
|
||||
float[][] predicts = booster.predict(dtest);
|
||||
//predict leaf
|
||||
float[][] leafPredicts = booster.predict(dtest, 0, true);
|
||||
```
|
||||
177
doc/jvm/java_intro.rst
Normal file
177
doc/jvm/java_intro.rst
Normal file
@ -0,0 +1,177 @@
|
||||
##################
|
||||
XGBoost4J Java API
|
||||
##################
|
||||
This tutorial introduces Java API for XGBoost.
|
||||
|
||||
**************
|
||||
Data Interface
|
||||
**************
|
||||
Like the XGBoost python module, XGBoost4J uses ``DMatrix`` to handle data,
|
||||
libsvm txt format file, sparse matrix in CSR/CSC format, and dense matrix is
|
||||
supported.
|
||||
|
||||
* The first step is to import ``DMatrix``:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
import org.dmlc.xgboost4j.DMatrix;
|
||||
|
||||
* Use ``DMatrix`` constructor to load data from a libsvm text format file:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
DMatrix dmat = new DMatrix("train.svm.txt");
|
||||
|
||||
* Pass arrays to ``DMatrix`` constructor to load from sparse matrix.
|
||||
|
||||
Suppose we have a sparse matrix
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
1 0 2 0
|
||||
4 0 0 3
|
||||
3 1 2 0
|
||||
|
||||
We can express the sparse matrix in `Compressed Sparse Row (CSR) <https://en.wikipedia.org/wiki/Sparse_matrix#Compressed_sparse_row_(CSR,_CRS_or_Yale_format)>`_ format:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
long[] rowHeaders = new long[] {0,2,4,7};
|
||||
float[] data = new float[] {1f,2f,4f,3f,3f,1f,2f};
|
||||
int[] colIndex = new int[] {0,2,0,3,0,1,2};
|
||||
DMatrix dmat = new DMatrix(rowHeaders, colIndex, data, DMatrix.SparseType.CSR);
|
||||
|
||||
... or in `Compressed Sparse Column (CSC) <https://en.wikipedia.org/wiki/Sparse_matrix#Compressed_sparse_column_(CSC_or_CCS)>`_ format:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
long[] colHeaders = new long[] {0,3,4,6,7};
|
||||
float[] data = new float[] {1f,4f,3f,1f,2f,2f,3f};
|
||||
int[] rowIndex = new int[] {0,1,2,2,0,2,1};
|
||||
DMatrix dmat = new DMatrix(colHeaders, rowIndex, data, DMatrix.SparseType.CSC);
|
||||
|
||||
* You may also load your data from a dense matrix. Let's assume we have a matrix of form
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
1 2
|
||||
3 4
|
||||
5 6
|
||||
|
||||
Using `row-major layout <https://en.wikipedia.org/wiki/Row-_and_column-major_order>`_, we specify the dense matrix as follows:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
float[] data = new float[] {1f,2f,3f,4f,5f,6f};
|
||||
int nrow = 3;
|
||||
int ncol = 2;
|
||||
float missing = 0.0f;
|
||||
DMatrix dmat = new Matrix(data, nrow, ncol, missing);
|
||||
|
||||
* To set weight:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
float[] weights = new float[] {1f,2f,1f};
|
||||
dmat.setWeight(weights);
|
||||
|
||||
******************
|
||||
Setting Parameters
|
||||
******************
|
||||
* In XGBoost4J any ``Iterable<Entry<String, Object>>`` object could be used as parameters.
|
||||
|
||||
* To set parameters, for non-multiple value params, you can simply use entrySet of an Map:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
Map<String, Object> paramMap = new HashMap<>() {
|
||||
{
|
||||
put("eta", 1.0);
|
||||
put("max_depth", 2);
|
||||
put("silent", 1);
|
||||
put("objective", "binary:logistic");
|
||||
put("eval_metric", "logloss");
|
||||
}
|
||||
};
|
||||
Iterable<Entry<String, Object>> params = paramMap.entrySet();
|
||||
|
||||
* for the situation that multiple values with same param key, List<Entry<String, Object>> would be a good choice, e.g. :
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
List<Entry<String, Object>> params = new ArrayList<Entry<String, Object>>() {
|
||||
{
|
||||
add(new SimpleEntry<String, Object>("eta", 1.0));
|
||||
add(new SimpleEntry<String, Object>("max_depth", 2.0));
|
||||
add(new SimpleEntry<String, Object>("silent", 1));
|
||||
add(new SimpleEntry<String, Object>("objective", "binary:logistic"));
|
||||
}
|
||||
};
|
||||
|
||||
**************
|
||||
Training Model
|
||||
**************
|
||||
With parameters and data, you are able to train a booster model.
|
||||
|
||||
* Import ``Trainer`` and ``Booster``:
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
import org.dmlc.xgboost4j.Booster;
|
||||
import org.dmlc.xgboost4j.util.Trainer;
|
||||
|
||||
* Training
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
DMatrix trainMat = new DMatrix("train.svm.txt");
|
||||
DMatrix validMat = new DMatrix("valid.svm.txt");
|
||||
//specify a watchList to see the performance
|
||||
//any Iterable<Entry<String, DMatrix>> object could be used as watchList
|
||||
List<Entry<String, DMatrix>> watchs = new ArrayList<>();
|
||||
watchs.add(new SimpleEntry<>("train", trainMat));
|
||||
watchs.add(new SimpleEntry<>("test", testMat));
|
||||
int round = 2;
|
||||
Booster booster = Trainer.train(params, trainMat, round, watchs, null, null);
|
||||
|
||||
* Saving model
|
||||
|
||||
After training, you can save model and dump it out.
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
booster.saveModel("model.bin");
|
||||
|
||||
* Dump Model and Feature Map
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
booster.dumpModel("modelInfo.txt", false)
|
||||
//dump with featureMap
|
||||
booster.dumpModel("modelInfo.txt", "featureMap.txt", false)
|
||||
|
||||
* Load a model
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
Params param = new Params() {
|
||||
{
|
||||
put("silent", 1);
|
||||
put("nthread", 6);
|
||||
}
|
||||
};
|
||||
Booster booster = new Booster(param, "model.bin");
|
||||
|
||||
**********
|
||||
Prediction
|
||||
**********
|
||||
After training and loading a model, you can use it to make prediction for other data. The result will be a two-dimension float array ``(nsample, nclass)``; for ``predictLeaf()``, the result would be of shape ``(nsample, nclass*ntrees)``.
|
||||
|
||||
.. code-block:: java
|
||||
|
||||
DMatrix dtest = new DMatrix("test.svm.txt");
|
||||
//predict
|
||||
float[][] predicts = booster.predict(dtest);
|
||||
//predict leaf
|
||||
float[][] leafPredicts = booster.predict(dtest, 0, true);
|
||||
|
||||
@ -1,187 +0,0 @@
|
||||
---
|
||||
layout: post
|
||||
title: XGBoost4J: Portable Distributed Tree Boosting in DataFlow
|
||||
date: 2016-03-15 12:00:00
|
||||
author: Nan Zhu, Tianqi Chen
|
||||
comments: true
|
||||
---
|
||||
|
||||
## Introduction
|
||||
[XGBoost](https://github.com/dmlc/xgboost) is a library designed and optimized for tree boosting. Gradient boosting trees model is originally proposed by Friedman et al. By embracing multi-threads and introducing regularization, XGBoost delivers higher computational power and more accurate prediction. **More than half of the winning solutions in machine learning challenges** hosted at Kaggle adopt XGBoost ([Incomplete list](https://github.com/dmlc/xgboost/tree/master/demo#machine-learning-challenge-winning-solutions)).
|
||||
XGBoost has provided native interfaces for C++, R, python, Julia and Java users.
|
||||
It is used by both [data exploration and production scenarios](https://github.com/dmlc/xgboost/tree/master/demo#usecases) to solve real world machine learning problems.
|
||||
|
||||
The distributed XGBoost is described in the [recently published paper](http://arxiv.org/abs/1603.02754).
|
||||
In short, the XGBoost system runs magnitudes faster than existing alternatives of distributed ML,
|
||||
and uses far fewer resources. The reader is more than welcomed to refer to the paper for more details.
|
||||
|
||||
Despite the current great success, one of our ultimate goals is to make XGBoost even more available for all production scenario.
|
||||
Programming languages and data processing/storage systems based on Java Virtual Machine (JVM) play the significant roles in the BigData ecosystem. [Hadoop](http://hadoop.apache.org/), [Spark](http://spark.apache.org/) and more recently introduced [Flink](http://flink.apache.org/) are very useful solutions to general large-scale data processing.
|
||||
|
||||
On the other side, the emerging demands of machine learning and deep learning
|
||||
inspires many excellent machine learning libraries.
|
||||
Many of these machine learning libraries(e.g. [XGBoost](https://github.com/dmlc/xgboost)/[MxNet](https://github.com/dmlc/mxnet))
|
||||
requires new computation abstraction and native support (e.g. C++ for GPU computing).
|
||||
They are also often [much more efficient](http://arxiv.org/abs/1603.02754).
|
||||
|
||||
The gap between the implementation fundamentals of the general data processing frameworks and the more specific machine learning libraries/systems prohibits the smooth connection between these two types of systems, thus brings unnecessary inconvenience to the end user. The common workflow to the user is to utilize the systems like Spark/Flink to preprocess/clean data, pass the results to machine learning systems like [XGBoost](https://github.com/dmlc/xgboost)/[MxNet](https://github.com/dmlc/mxnet)) via the file systems and then conduct the following machine learning phase. This process jumping across two types of systems creates certain inconvenience for the users and brings additional overhead to the operators of the infrastructure.
|
||||
|
||||
We want best of both worlds, so we can use the data processing frameworks like Spark and Flink together with
|
||||
the best distributed machine learning solutions.
|
||||
To resolve the situation, we introduce the new-brewed [XGBoost4J](https://github.com/dmlc/xgboost/tree/master/jvm-packages),
|
||||
<b>XGBoost</b> for <b>J</b>VM Platform. We aim to provide the clean Java/Scala APIs and the integration with the most popular data processing systems developed in JVM-based languages.
|
||||
|
||||
## Unix Philosophy in Machine Learning
|
||||
|
||||
XGBoost and XGBoost4J adopts Unix Philosophy.
|
||||
XGBoost **does its best in one thing -- tree boosting** and is **being designed to work with other systems**.
|
||||
We strongly believe that machine learning solution should not be restricted to certain language or certain platform.
|
||||
|
||||
Specifically, users will be able to use distributed XGBoost in both Spark and Flink, and possibly more frameworks in Future.
|
||||
We have made the API in a portable way so it **can be easily ported to other Dataflow frameworks provided by the Cloud**.
|
||||
XGBoost4J shares its core with other XGBoost libraries, which means data scientists can use R/python
|
||||
read and visualize the model trained distributedly.
|
||||
It also means that user can start with single machine version for exploration,
|
||||
which already can handle hundreds of million examples.
|
||||
|
||||
## System Overview
|
||||
|
||||
In the following Figure, we describe the overall architecture of XGBoost4J. XGBoost4J provides the Java/Scala API calling the core functionality of XGBoost library. Most importantly, it not only supports the single-machine model training, but also provides an abstraction layer which masks the difference of the underlying data processing engines and scales training to the distributed servers.
|
||||
|
||||

|
||||
|
||||
|
||||
By calling the XGBoost4J API, users can scale the model training to the cluster. XGBoost4J calls the running instance of XGBoost worker in Spark/Flink task and run them across the cluster. The communication among the distributed model training tasks and the XGBoost4J runtime environment go through [Rabit] (https://github.com/dmlc/rabit).
|
||||
|
||||
With the abstraction of XGBoost4J, users can build an unified data analytic application ranging from Extract-Transform-Loading, data exploration, machine learning model training and the final data product service. The following figure illustrate an example application built on top of Apache Spark. The application seamlessly embeds XGBoost into the processing pipeline and exchange data with other Spark-based processing phase through Spark's distributed memory layer.
|
||||
|
||||

|
||||
|
||||
|
||||
## Single-machine Training Walk-through
|
||||
|
||||
In this section, we will work through the APIs of XGBoost4J by examples.
|
||||
We will be using scala for demonstration, but we also have a complete API for java users.
|
||||
|
||||
To start the model training and evaluation, we need to prepare the training and test set:
|
||||
|
||||
```scala
|
||||
val trainMax = new DMatrix("../../demo/data/agaricus.txt.train")
|
||||
val testMax = new DMatrix("../../demo/data/agaricus.txt.test")
|
||||
```
|
||||
|
||||
After preparing the data, we can train our model:
|
||||
|
||||
```scala
|
||||
val params = new mutable.HashMap[String, Any]()
|
||||
params += "eta" -> 1.0
|
||||
params += "max_depth" -> 2
|
||||
params += "silent" -> 1
|
||||
params += "objective" -> "binary:logistic"
|
||||
|
||||
val watches = new mutable.HashMap[String, DMatrix]
|
||||
watches += "train" -> trainMax
|
||||
watches += "test" -> testMax
|
||||
|
||||
val round = 2
|
||||
// train a model
|
||||
val booster = XGBoost.train(trainMax, params.toMap, round, watches.toMap)
|
||||
```
|
||||
|
||||
We then evaluate our model:
|
||||
|
||||
```scala
|
||||
val predicts = booster.predict(testMax)
|
||||
```
|
||||
|
||||
`predict` can output the predict results and you can define a customized evaluation method to derive your own metrics (see the example in ([Customized Evaluation Metric in Java](https://github.com/dmlc/xgboost/blob/master/jvm-packages/xgboost4j-example/src/main/java/ml/dmlc/xgboost4j/java/example/CustomObjective.java), [Customized Evaluation Metric in Scala] (https://github.com/dmlc/xgboost/blob/master/jvm-packages/xgboost4j-example/src/main/scala/ml/dmlc/xgboost4j/scala/example/CustomObjective.scala)).
|
||||
|
||||
|
||||
## Distributed Model Training with Distributed Dataflow Frameworks
|
||||
|
||||
The most exciting part in this XGBoost4J release is the integration with the Distributed Dataflow Framework. The most popular data processing frameworks fall into this category, e.g. [Apache Spark](http://spark.apache.org/), [Apache Flink] (http://flink.apache.org/), etc. In this part, we will walk through the steps to build the unified data analytic applications containing data preprocessing and distributed model training with Spark and Flink. (currently, we only provide Scala API for the integration with Spark and Flink)
|
||||
|
||||
Similar to the single-machine training, we need to prepare the training and test dataset.
|
||||
|
||||
### Spark Example
|
||||
|
||||
In Spark, the dataset is represented as the [Resilient Distributed Dataset (RDD)](http://spark.apache.org/docs/latest/programming-guide.html#resilient-distributed-datasets-rdds), we can utilize the Spark-distributed tools to parse libSVM file and wrap it as the RDD:
|
||||
|
||||
```scala
|
||||
val trainRDD = MLUtils.loadLibSVMFile(sc, inputTrainPath).repartition(args(1).toInt)
|
||||
```
|
||||
|
||||
We move forward to train the models:
|
||||
|
||||
```scala
|
||||
val xgboostModel = XGBoost.train(trainRDD, paramMap, numRound, numWorkers)
|
||||
|
||||
```
|
||||
|
||||
The next step is to evaluate the model, you can either predict in local side or in a distributed fashion
|
||||
|
||||
|
||||
```scala
|
||||
// testSet is an RDD containing testset data represented as
|
||||
// org.apache.spark.mllib.regression.LabeledPoint
|
||||
val testSet = MLUtils.loadLibSVMFile(sc, inputTestPath)
|
||||
|
||||
// local prediction
|
||||
// import methods in DataUtils to convert Iterator[org.apache.spark.mllib.regression.LabeledPoint]
|
||||
// to Iterator[ml.dmlc.xgboost4j.LabeledPoint] in automatic
|
||||
import DataUtils._
|
||||
xgboostModel.predict(new DMatrix(testSet.collect().iterator)
|
||||
|
||||
// distributed prediction
|
||||
xgboostModel.predict(testSet)
|
||||
|
||||
```
|
||||
### Flink example
|
||||
|
||||
In Flink, we represent training data as Flink's [DataSet](https://ci.apache.org/projects/flink/flink-docs-master/apis/batch/index.html)
|
||||
|
||||
```scala
|
||||
val trainData = MLUtils.readLibSVM(env, "/path/to/data/agaricus.txt.train")
|
||||
```
|
||||
|
||||
Model Training can be done as follows
|
||||
|
||||
```scala
|
||||
val xgboostModel = XGBoost.train(trainData, paramMap, round)
|
||||
|
||||
```
|
||||
|
||||
Training and prediction.
|
||||
|
||||
```scala
|
||||
// testData is a Dataset containing testset data represented as
|
||||
// org.apache.flink.ml.math.Vector.LabeledVector
|
||||
val testData = MLUtils.readLibSVM(env, "/path/to/data/agaricus.txt.test")
|
||||
|
||||
// local prediction
|
||||
xgboostModel.predict(testData.collect().iterator)
|
||||
|
||||
// distributed prediction
|
||||
xgboostModel.predict(testData.map{x => x.vector})
|
||||
```
|
||||
|
||||
## Road Map
|
||||
|
||||
It is the first release of XGBoost4J package, we are actively move forward for more charming features in the next release. You can watch our progress in [XGBoost4J Road Map](https://github.com/dmlc/xgboost/issues/935).
|
||||
|
||||
While we are trying our best to keep the minimum changes to the APIs, it is still subject to the incompatible changes.
|
||||
|
||||
## Further Readings
|
||||
|
||||
If you are interested in knowing more about XGBoost, you can find rich resources in
|
||||
|
||||
- [The github repository of XGBoost](https://github.com/dmlc/xgboost)
|
||||
- [The comprehensive documentation site for XGBoostl](http://xgboost.readthedocs.org/en/latest/index.html)
|
||||
- [An introduction to the gradient boosting model](http://xgboost.readthedocs.org/en/latest/model.html)
|
||||
- [Tutorials for the R package](xgboost.readthedocs.org/en/latest/R-package/index.html)
|
||||
- [Introduction of the Parameters](http://xgboost.readthedocs.org/en/latest/parameter.html)
|
||||
- [Awesome XGBoost, a curated list of examples, tutorials, blogs about XGBoost usecases](https://github.com/dmlc/xgboost/tree/master/demo)
|
||||
|
||||
## Acknowledgements
|
||||
|
||||
We would like to send many thanks to [Zixuan Huang](https://github.com/yanqingmen), the early developer of XGBoost for Java (XGBoost for Java).
|
||||
@ -1,139 +0,0 @@
|
||||
## Introduction
|
||||
|
||||
On March 2016, we released the first version of [XGBoost4J](http://dmlc.ml/2016/03/14/xgboost4j-portable-distributed-xgboost-in-spark-flink-and-dataflow.html), which is a set of packages providing Java/Scala interfaces of XGBoost and the integration with prevalent JVM-based distributed data processing platforms, like Spark/Flink.
|
||||
|
||||
The integrations with Spark/Flink, a.k.a. <b>XGBoost4J-Spark</b> and <b>XGBoost-Flink</b>, receive the tremendous positive feedbacks from the community. It enables users to build a unified pipeline, embedding XGBoost into the data processing system based on the widely-deployed frameworks like Spark. The following figure shows the general architecture of such a pipeline with the first version of <b>XGBoost4J-Spark</b>, where the data processing is based on the low-level [Resilient Distributed Dataset (RDD)](http://spark.apache.org/docs/latest/programming-guide.html#resilient-distributed-datasets-rdds) abstraction.
|
||||
|
||||

|
||||
|
||||
In the last months, we have a lot of communication with the users and gain the deeper understanding of the users' latest usage scenario and requirements:
|
||||
|
||||
* XGBoost keeps gaining more and more deployments in the production environment and the adoption in machine learning competitions [Link](http://datascience.la/xgboost-workshop-and-meetup-talk-with-tianqi-chen/).
|
||||
|
||||
* While Spark is still the mainstream data processing tool in most of scenarios, more and more users are porting their RDD-based Spark programs to [DataFrame/Dataset APIs](http://spark.apache.org/docs/latest/sql-programming-guide.html) for the well-designed interfaces to manipulate structured data and the [significant performance improvement](https://databricks.com/blog/2016/07/26/introducing-apache-spark-2-0.html).
|
||||
|
||||
* Spark itself has presented a clear roadmap that DataFrame/Dataset would be the base of the latest and future features, e.g. latest version of [ML pipeline](http://spark.apache.org/docs/latest/ml-guide.html) and [Structured Streaming](http://spark.apache.org/docs/latest/structured-streaming-programming-guide.html).
|
||||
|
||||
Based on these feedbacks from the users, we observe a gap between the original RDD-based XGBoost4J-Spark and the users' latest usage scenario as well as the future direction of Spark ecosystem. To fill this gap, we start working on the <b><i>integration of XGBoost and Spark's DataFrame/Dataset abstraction</i></b> in September. In this blog, we will introduce <b>the latest version of XGBoost4J-Spark</b> which allows the user to work with DataFrame/Dataset directly and embed XGBoost to Spark's ML pipeline seamlessly.
|
||||
|
||||
|
||||
## A Full Integration of XGBoost and DataFrame/Dataset
|
||||
|
||||
The following figure illustrates the new pipeline architecture with the latest XGBoost4J-Spark.
|
||||
|
||||

|
||||
|
||||
Being different with the previous version, users are able to use both low- and high-level memory abstraction in Spark, i.e. RDD and DataFrame/Dataset. The DataFrame/Dataset abstraction grants the user to manipulate structured datasets and utilize the built-in routines in Spark or User Defined Functions (UDF) to explore the value distribution in columns before they feed data into the machine learning phase in the pipeline. In the following example, the structured sales records can be saved in a JSON file, parsed as DataFrame through Spark's API and feed to train XGBoost model in two lines of Scala code.
|
||||
|
||||
```scala
|
||||
// load sales records saved in json files
|
||||
val salesDF = spark.read.json("sales.json")
|
||||
// call XGBoost API to train with the DataFrame-represented training set
|
||||
val xgboostModel = XGBoost.trainWithDataFrame(
|
||||
salesDF, paramMap, numRound, nWorkers, useExternalMemory)
|
||||
```
|
||||
|
||||
By integrating with DataFrame/Dataset, XGBoost4J-Spark not only enables users to call DataFrame/Dataset APIs directly but also make DataFrame/Dataset-based Spark features available to XGBoost users, e.g. ML Package.
|
||||
|
||||
### Integration with ML Package
|
||||
|
||||
ML package of Spark provides a set of convenient tools for feature extraction/transformation/selection. Additionally, with the model selection tool in ML package, users can select the best model through an automatic parameter searching process which is defined with through ML package APIs. After integrating with DataFrame/Dataset abstraction, these charming features in ML package are also available to XGBoost users.
|
||||
|
||||
#### Feature Extraction/Transformation/Selection
|
||||
|
||||
The following example shows a feature transformer which converts the string-typed storeType feature to the numeric storeTypeIndex. The transformed DataFrame is then fed to train XGBoost model.
|
||||
|
||||
```scala
|
||||
import org.apache.spark.ml.feature.StringIndexer
|
||||
|
||||
// load sales records saved in json files
|
||||
val salesDF = spark.read.json("sales.json")
|
||||
|
||||
// transform the string-represented storeType feature to numeric storeTypeIndex
|
||||
val indexer = new StringIndexer()
|
||||
.setInputCol("storeType")
|
||||
.setOutputCol("storeTypeIndex")
|
||||
// drop the extra column
|
||||
val indexed = indexer.fit(salesDF).transform(df).drop("storeType")
|
||||
|
||||
// use the transformed dataframe as training dataset
|
||||
val xgboostModel = XGBoost.trainWithDataFrame(
|
||||
indexed, paramMap, numRound, nWorkers, useExternalMemory)
|
||||
```
|
||||
|
||||
#### Pipelining
|
||||
|
||||
Spark ML package allows the user to build a complete pipeline from feature extraction/transformation/selection to model training. We integrate XGBoost with ML package and make it feasible to embed XGBoost into such a pipeline seamlessly. The following example shows how to build such a pipeline consisting of feature transformers and the XGBoost estimator.
|
||||
|
||||
```scala
|
||||
import org.apache.spark.ml.feature.StringIndexer
|
||||
|
||||
// load sales records saved in json files
|
||||
val salesDF = spark.read.json("sales.json")
|
||||
|
||||
// transform the string-represented storeType feature to numeric storeTypeIndex
|
||||
val indexer = new StringIndexer()
|
||||
.setInputCol("storeType")
|
||||
.setOutputCol("storeTypeIndex")
|
||||
|
||||
// assemble the columns in dataframe into a vector
|
||||
val vectorAssembler = new VectorAssembler()
|
||||
.setInputCols(Array("storeId", "storeTypeIndex", ...))
|
||||
.setOutputCol("features")
|
||||
|
||||
// construct the pipeline
|
||||
val pipeline = new Pipeline().setStages(
|
||||
Array(storeTypeIndexer, ..., vectorAssembler, new XGBoostEstimator(Map[String, Any]("num_rounds" -> 100)))
|
||||
|
||||
// use the transformed dataframe as training dataset
|
||||
val xgboostModel = pipeline.fit(salesDF)
|
||||
|
||||
// predict with the trained model
|
||||
val salesTestDF = spark.read.json("sales_test.json")
|
||||
val salesRecordsWithPred = xgboostModel.transform(salesTestDF)
|
||||
|
||||
```
|
||||
|
||||
#### Model Selection
|
||||
|
||||
The most critical operation to maximize the power of XGBoost is to select the optimal parameters for the model. Tuning parameters manually is a tedious and labor-consuming process. With the latest version of XGBoost4J-Spark, we can utilize the Spark model selecting tool to automate this process. The following example shows the code snippet utilizing [TrainValidationSplit](http://spark.apache.org/docs/latest/api/scala/index.html#org.apache.spark.ml.tuning.TrainValidationSplit) and [RegressionEvaluator](http://spark.apache.org/docs/latest/api/scala/index.html#org.apache.spark.ml.evaluation.RegressionEvaluator) to search the optimal combination of two XGBoost parameters, [max_depth and eta] (https://github.com/dmlc/xgboost/blob/master/doc/parameter.md). The model producing the minimum cost function value defined by RegressionEvaluator is selected and used to generate the prediction for the test set.
|
||||
|
||||
```scala
|
||||
// create XGBoostEstimator
|
||||
val xgbEstimator = new XGBoostEstimator(xgboostParam).setFeaturesCol("features").
|
||||
setLabelCol("sales")
|
||||
val paramGrid = new ParamGridBuilder()
|
||||
.addGrid(xgbEstimator.maxDepth, Array(5, 6))
|
||||
.addGrid(xgbEstimator.eta, Array(0.1, 0.4))
|
||||
.build()
|
||||
val tv = new TrainValidationSplit()
|
||||
.setEstimator(xgbEstimator)
|
||||
.setEvaluator(new RegressionEvaluator().setLabelCol("sales"))
|
||||
.setEstimatorParamMaps(paramGrid)
|
||||
.setTrainRatio(0.8)
|
||||
val salesTestDF = spark.read.json("sales_test.json")
|
||||
val salesRecordsWithPred = xgboostModel.transform(salesTestDF)
|
||||
```
|
||||
|
||||
## Summary
|
||||
|
||||
Through the latest XGBoost4J-Spark, XGBoost users can build a more efficient data processing pipeline which works with DataFrame/Dataset APIs to handle the structured data with the excellent performance, and simultaneously embrace the powerful XGBoost to explore the insights from the dataset and transform this insight into action. Additionally, XGBoost4J-Spark seamlessly connect XGBoost with Spark ML package which makes the job of feature extraction/transformation/selection and parameter model much easier than before.
|
||||
|
||||
The latest version of XGBoost4J-Spark has been available in the [GitHub Repository] (https://github.com/dmlc/xgboost), and the latest API docs are in [here](http://xgboost.readthedocs.io/en/latest/jvm/index.html).
|
||||
|
||||
|
||||
## Portable Machine Learning Systems
|
||||
|
||||
XGBoost is one of the projects incubated by [Distributed Machine Learning Community (DMLC)](http://dmlc.ml/), which also creates several other popular projects on machine learning systems ([Link](https://github.com/dmlc/)), e.g. one of the most popular deep learning frameworks, [MXNet](http://mxnet.io/). We strongly believe that machine learning solution should not be restricted to certain language or certain platform. We realize this design philosophy in several projects, like XGBoost and MXNet. We are willing to see more contributions from the community in this direction.
|
||||
|
||||
|
||||
## Further Readings
|
||||
|
||||
If you are interested in knowing more about XGBoost, you can find rich resources in
|
||||
|
||||
- [The github repository of XGBoost](https://github.com/dmlc/xgboost)
|
||||
- [The comprehensive documentation site for XGBoostl](http://xgboost.readthedocs.org/en/latest/index.html)
|
||||
- [An introduction to the gradient boosting model](http://xgboost.readthedocs.org/en/latest/model.html)
|
||||
- [Tutorials for the R package](xgboost.readthedocs.org/en/latest/R-package/index.html)
|
||||
- [Introduction of the Parameters](http://xgboost.readthedocs.org/en/latest/parameter.html)
|
||||
- [Awesome XGBoost, a curated list of examples, tutorials, blogs about XGBoost usecases](https://github.com/dmlc/xgboost/tree/master/demo)
|
||||
241
doc/model.md
241
doc/model.md
@ -1,241 +0,0 @@
|
||||
Introduction to Boosted Trees
|
||||
=============================
|
||||
XGBoost is short for "Extreme Gradient Boosting", where the term "Gradient Boosting" is proposed in the paper _Greedy Function Approximation: A Gradient Boosting Machine_, by Friedman.
|
||||
XGBoost is based on this original model.
|
||||
This is a tutorial on gradient boosted trees, and most of the content is based on these [slides](http://homes.cs.washington.edu/~tqchen/pdf/BoostedTree.pdf) by the author of xgboost.
|
||||
|
||||
The GBM (boosted trees) has been around for really a while, and there are a lot of materials on the topic.
|
||||
This tutorial tries to explain boosted trees in a self-contained and principled way using the elements of supervised learning.
|
||||
We think this explanation is cleaner, more formal, and motivates the variant used in xgboost.
|
||||
|
||||
Elements of Supervised Learning
|
||||
-------------------------------
|
||||
XGBoost is used for supervised learning problems, where we use the training data (with multiple features) ``$ x_i $`` to predict a target variable ``$ y_i $``.
|
||||
Before we dive into trees, let us start by reviewing the basic elements in supervised learning.
|
||||
|
||||
### Model and Parameters
|
||||
The ***model*** in supervised learning usually refers to the mathematical structure of how to make the prediction ``$ y_i $`` given ``$ x_i $``.
|
||||
For example, a common model is a *linear model*, where the prediction is given by ``$ \hat{y}_i = \sum_j \theta_j x_{ij} $``, a linear combination of weighted input features.
|
||||
The prediction value can have different interpretations, depending on the task, i.e., regression or classification.
|
||||
For example, it can be logistic transformed to get the probability of positive class in logistic regression, and it can also be used as a ranking score when we want to rank the outputs.
|
||||
|
||||
The ***parameters*** are the undetermined part that we need to learn from data. In linear regression problems, the parameters are the coefficients ``$ \theta $``.
|
||||
Usually we will use ``$ \theta $`` to denote the parameters (there are many parameters in a model, our definition here is sloppy).
|
||||
|
||||
### Objective Function : Training Loss + Regularization
|
||||
|
||||
Based on different understandings of ``$ y_i $`` we can have different problems, such as regression, classification, ordering, etc.
|
||||
We need to find a way to find the best parameters given the training data. In order to do so, we need to define a so-called ***objective function***,
|
||||
to measure the performance of the model given a certain set of parameters.
|
||||
|
||||
A very important fact about objective functions is they ***must always*** contain two parts: training loss and regularization.
|
||||
|
||||
```math
|
||||
\text{obj}(\theta) = L(\theta) + \Omega(\theta)
|
||||
```
|
||||
|
||||
where ``$ L $`` is the training loss function, and ``$ \Omega $`` is the regularization term. The training loss measures how *predictive* our model is on training data.
|
||||
For example, a commonly used training loss is mean squared error.
|
||||
|
||||
```math
|
||||
L(\theta) = \sum_i (y_i-\hat{y}_i)^2
|
||||
```
|
||||
Another commonly used loss function is logistic loss for logistic regression
|
||||
|
||||
```math
|
||||
L(\theta) = \sum_i[ y_i\ln (1+e^{-\hat{y}_i}) + (1-y_i)\ln (1+e^{\hat{y}_i})]
|
||||
```
|
||||
|
||||
The ***regularization term*** is what people usually forget to add. The regularization term controls the complexity of the model, which helps us to avoid overfitting.
|
||||
This sounds a bit abstract, so let us consider the following problem in the following picture. You are asked to *fit* visually a step function given the input data points
|
||||
on the upper left corner of the image.
|
||||
Which solution among the three do you think is the best fit?
|
||||
|
||||
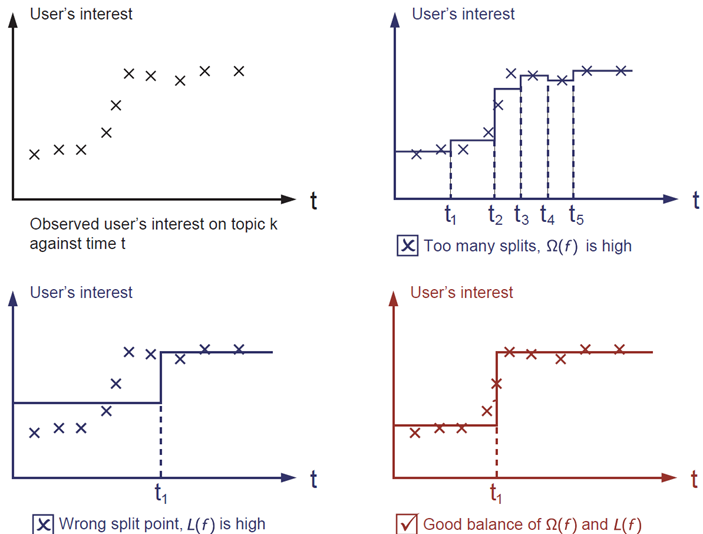
|
||||
|
||||
The correct answer is marked in red. Please consider if this visually seems a reasonable fit to you. The general principle is we want both a ***simple*** and ***predictive*** model.
|
||||
The tradeoff between the two is also referred as bias-variance tradeoff in machine learning.
|
||||
|
||||
|
||||
### Why introduce the general principle?
|
||||
The elements introduced above form the basic elements of supervised learning, and they are naturally the building blocks of machine learning toolkits.
|
||||
For example, you should be able to describe the differences and commonalities between boosted trees and random forests.
|
||||
Understanding the process in a formalized way also helps us to understand the objective that we are learning and the reason behind the heuristics such as
|
||||
pruning and smoothing.
|
||||
|
||||
Tree Ensemble
|
||||
-------------
|
||||
Now that we have introduced the elements of supervised learning, let us get started with real trees.
|
||||
To begin with, let us first learn about the ***model*** of xgboost: tree ensembles.
|
||||
The tree ensemble model is a set of classification and regression trees (CART). Here's a simple example of a CART
|
||||
that classifies whether someone will like computer games.
|
||||
|
||||
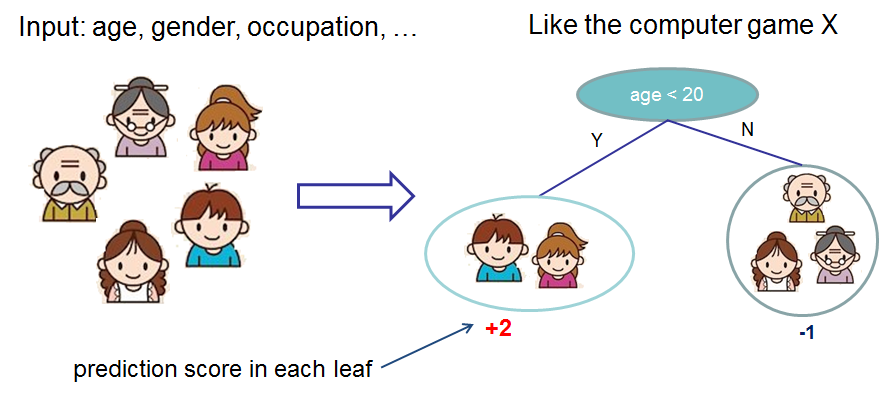
|
||||
|
||||
We classify the members of a family into different leaves, and assign them the score on the corresponding leaf.
|
||||
A CART is a bit different from decision trees, where the leaf only contains decision values. In CART, a real score
|
||||
is associated with each of the leaves, which gives us richer interpretations that go beyond classification.
|
||||
This also makes the unified optimization step easier, as we will see in a later part of this tutorial.
|
||||
|
||||
Usually, a single tree is not strong enough to be used in practice. What is actually used is the so-called
|
||||
tree ensemble model, which sums the prediction of multiple trees together.
|
||||
|
||||
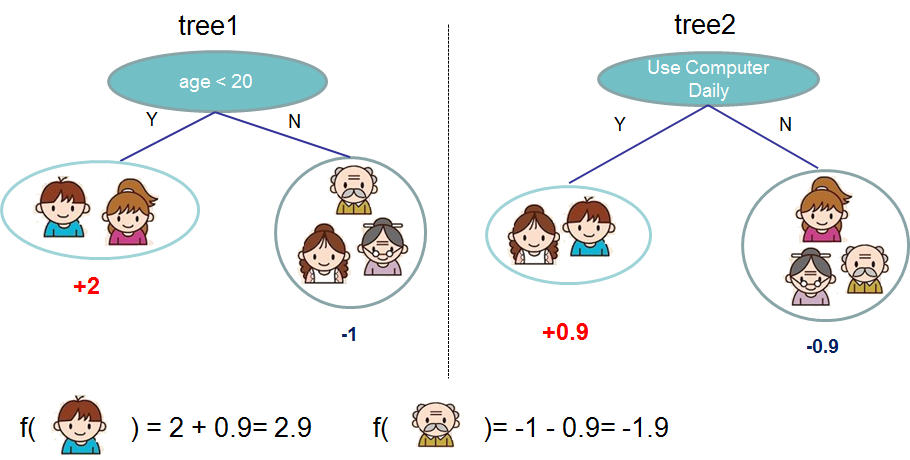
|
||||
|
||||
Here is an example of a tree ensemble of two trees. The prediction scores of each individual tree are summed up to get the final score.
|
||||
If you look at the example, an important fact is that the two trees try to *complement* each other.
|
||||
Mathematically, we can write our model in the form
|
||||
|
||||
```math
|
||||
\hat{y}_i = \sum_{k=1}^K f_k(x_i), f_k \in \mathcal{F}
|
||||
```
|
||||
|
||||
where ``$ K $`` is the number of trees, ``$ f $`` is a function in the functional space ``$ \mathcal{F} $``, and ``$ \mathcal{F} $`` is the set of all possible CARTs. Therefore our objective to optimize can be written as
|
||||
|
||||
```math
|
||||
\text{obj}(\theta) = \sum_i^n l(y_i, \hat{y}_i) + \sum_{k=1}^K \Omega(f_k)
|
||||
```
|
||||
Now here comes the question, what is the *model* for random forests? It is exactly tree ensembles! So random forests and boosted trees are not different in terms of model,
|
||||
the difference is how we train them. This means if you write a predictive service of tree ensembles, you only need to write one of them and they should directly work
|
||||
for both random forests and boosted trees. One example of why elements of supervised learning rock.
|
||||
|
||||
Tree Boosting
|
||||
-------------
|
||||
After introducing the model, let us begin with the real training part. How should we learn the trees?
|
||||
The answer is, as is always for all supervised learning models: *define an objective function, and optimize it*!
|
||||
|
||||
Assume we have the following objective function (remember it always needs to contain training loss and regularization)
|
||||
```math
|
||||
\text{obj} = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t)}) + \sum_{i=1}^t\Omega(f_i) \\
|
||||
```
|
||||
|
||||
### Additive Training
|
||||
|
||||
First thing we want to ask is what are the ***parameters*** of trees?
|
||||
You can find that what we need to learn are those functions ``$f_i$``, with each containing the structure
|
||||
of the tree and the leaf scores. This is much harder than traditional optimization problem where you can take the gradient and go.
|
||||
It is not easy to train all the trees at once.
|
||||
Instead, we use an additive strategy: fix what we have learned, and add one new tree at a time.
|
||||
We write the prediction value at step ``$t$`` as ``$ \hat{y}_i^{(t)}$``, so we have
|
||||
|
||||
```math
|
||||
\hat{y}_i^{(0)} &= 0\\
|
||||
\hat{y}_i^{(1)} &= f_1(x_i) = \hat{y}_i^{(0)} + f_1(x_i)\\
|
||||
\hat{y}_i^{(2)} &= f_1(x_i) + f_2(x_i)= \hat{y}_i^{(1)} + f_2(x_i)\\
|
||||
&\dots\\
|
||||
\hat{y}_i^{(t)} &= \sum_{k=1}^t f_k(x_i)= \hat{y}_i^{(t-1)} + f_t(x_i)
|
||||
```
|
||||
|
||||
It remains to ask, which tree do we want at each step? A natural thing is to add the one that optimizes our objective.
|
||||
|
||||
```math
|
||||
\text{obj}^{(t)} & = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t)}) + \sum_{i=1}^t\Omega(f_i) \\
|
||||
& = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t-1)} + f_t(x_i)) + \Omega(f_t) + constant
|
||||
```
|
||||
|
||||
If we consider using MSE as our loss function, it becomes the following form.
|
||||
|
||||
```math
|
||||
\text{obj}^{(t)} & = \sum_{i=1}^n (y_i - (\hat{y}_i^{(t-1)} + f_t(x_i)))^2 + \sum_{i=1}^t\Omega(f_i) \\
|
||||
& = \sum_{i=1}^n [2(\hat{y}_i^{(t-1)} - y_i)f_t(x_i) + f_t(x_i)^2] + \Omega(f_t) + constant
|
||||
```
|
||||
|
||||
The form of MSE is friendly, with a first order term (usually called the residual) and a quadratic term.
|
||||
For other losses of interest (for example, logistic loss), it is not so easy to get such a nice form.
|
||||
So in the general case, we take the Taylor expansion of the loss function up to the second order
|
||||
|
||||
```math
|
||||
\text{obj}^{(t)} = \sum_{i=1}^n [l(y_i, \hat{y}_i^{(t-1)}) + g_i f_t(x_i) + \frac{1}{2} h_i f_t^2(x_i)] + \Omega(f_t) + constant
|
||||
```
|
||||
where the ``$g_i$`` and ``$h_i$`` are defined as
|
||||
|
||||
```math
|
||||
g_i &= \partial_{\hat{y}_i^{(t-1)}} l(y_i, \hat{y}_i^{(t-1)})\\
|
||||
h_i &= \partial_{\hat{y}_i^{(t-1)}}^2 l(y_i, \hat{y}_i^{(t-1)})
|
||||
```
|
||||
|
||||
After we remove all the constants, the specific objective at step ``$t$`` becomes
|
||||
|
||||
```math
|
||||
\sum_{i=1}^n [g_i f_t(x_i) + \frac{1}{2} h_i f_t^2(x_i)] + \Omega(f_t)
|
||||
```
|
||||
|
||||
This becomes our optimization goal for the new tree. One important advantage of this definition is that
|
||||
it only depends on ``$g_i$`` and ``$h_i$``. This is how xgboost can support custom loss functions.
|
||||
We can optimize every loss function, including logistic regression and weighted logistic regression, using exactly
|
||||
the same solver that takes ``$g_i$`` and ``$h_i$`` as input!
|
||||
|
||||
### Model Complexity
|
||||
We have introduced the training step, but wait, there is one important thing, the ***regularization***!
|
||||
We need to define the complexity of the tree ``$\Omega(f)$``. In order to do so, let us first refine the definition of the tree ``$ f(x) $`` as
|
||||
|
||||
```math
|
||||
f_t(x) = w_{q(x)}, w \in R^T, q:R^d\rightarrow \{1,2,\cdots,T\} .
|
||||
```
|
||||
|
||||
Here ``$ w $`` is the vector of scores on leaves, ``$ q $`` is a function assigning each data point to the corresponding leaf, and ``$ T $`` is the number of leaves.
|
||||
In XGBoost, we define the complexity as
|
||||
|
||||
```math
|
||||
\Omega(f) = \gamma T + \frac{1}{2}\lambda \sum_{j=1}^T w_j^2
|
||||
```
|
||||
Of course there is more than one way to define the complexity, but this specific one works well in practice. The regularization is one part most tree packages treat
|
||||
less carefully, or simply ignore. This was because the traditional treatment of tree learning only emphasized improving impurity, while the complexity control was left to heuristics.
|
||||
By defining it formally, we can get a better idea of what we are learning, and yes it works well in practice.
|
||||
|
||||
### The Structure Score
|
||||
|
||||
Here is the magical part of the derivation. After reformalizing the tree model, we can write the objective value with the ``$ t$``-th tree as:
|
||||
|
||||
```math
|
||||
\text{obj}^{(t)} &\approx \sum_{i=1}^n [g_i w_{q(x_i)} + \frac{1}{2} h_i w_{q(x_i)}^2] + \gamma T + \frac{1}{2}\lambda \sum_{j=1}^T w_j^2\\
|
||||
&= \sum^T_{j=1} [(\sum_{i\in I_j} g_i) w_j + \frac{1}{2} (\sum_{i\in I_j} h_i + \lambda) w_j^2 ] + \gamma T
|
||||
```
|
||||
|
||||
where ``$ I_j = \{i|q(x_i)=j\} $`` is the set of indices of data points assigned to the ``$ j $``-th leaf.
|
||||
Notice that in the second line we have changed the index of the summation because all the data points on the same leaf get the same score.
|
||||
We could further compress the expression by defining ``$ G_j = \sum_{i\in I_j} g_i $`` and ``$ H_j = \sum_{i\in I_j} h_i $``:
|
||||
|
||||
```math
|
||||
\text{obj}^{(t)} = \sum^T_{j=1} [G_jw_j + \frac{1}{2} (H_j+\lambda) w_j^2] +\gamma T
|
||||
```
|
||||
|
||||
In this equation ``$ w_j $`` are independent with respect to each other, the form ``$ G_jw_j+\frac{1}{2}(H_j+\lambda)w_j^2 $`` is quadratic and the best ``$ w_j $`` for a given structure ``$q(x)$`` and the best objective reduction we can get is:
|
||||
|
||||
```math
|
||||
w_j^\ast = -\frac{G_j}{H_j+\lambda}\\
|
||||
\text{obj}^\ast = -\frac{1}{2} \sum_{j=1}^T \frac{G_j^2}{H_j+\lambda} + \gamma T
|
||||
```
|
||||
The last equation measures ***how good*** a tree structure ``$q(x)$`` is.
|
||||
|
||||
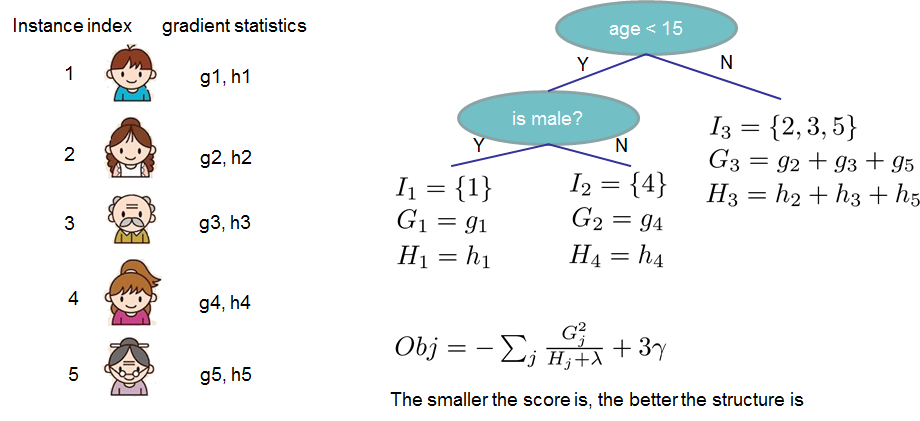
|
||||
|
||||
If all this sounds a bit complicated, let's take a look at the picture, and see how the scores can be calculated.
|
||||
Basically, for a given tree structure, we push the statistics ``$g_i$`` and ``$h_i$`` to the leaves they belong to,
|
||||
sum the statistics together, and use the formula to calculate how good the tree is.
|
||||
This score is like the impurity measure in a decision tree, except that it also takes the model complexity into account.
|
||||
|
||||
### Learn the tree structure
|
||||
Now that we have a way to measure how good a tree is, ideally we would enumerate all possible trees and pick the best one.
|
||||
In practice this is intractable, so we will try to optimize one level of the tree at a time.
|
||||
Specifically we try to split a leaf into two leaves, and the score it gains is
|
||||
|
||||
```math
|
||||
Gain = \frac{1}{2} \left[\frac{G_L^2}{H_L+\lambda}+\frac{G_R^2}{H_R+\lambda}-\frac{(G_L+G_R)^2}{H_L+H_R+\lambda}\right] - \gamma
|
||||
```
|
||||
This formula can be decomposed as 1) the score on the new left leaf 2) the score on the new right leaf 3) The score on the original leaf 4) regularization on the additional leaf.
|
||||
We can see an important fact here: if the gain is smaller than ``$\gamma$``, we would do better not to add that branch. This is exactly the ***pruning*** techniques in tree based
|
||||
models! By using the principles of supervised learning, we can naturally come up with the reason these techniques work :)
|
||||
|
||||
For real valued data, we usually want to search for an optimal split. To efficiently do so, we place all the instances in sorted order, like the following picture.
|
||||
|
||||
|
||||
A left to right scan is sufficient to calculate the structure score of all possible split solutions, and we can find the best split efficiently.
|
||||
|
||||
Final words on XGBoost
|
||||
----------------------
|
||||
Now that you understand what boosted trees are, you may ask, where is the introduction on [XGBoost](https://github.com/dmlc/xgboost)?
|
||||
XGBoost is exactly a tool motivated by the formal principle introduced in this tutorial!
|
||||
More importantly, it is developed with both deep consideration in terms of ***systems optimization*** and ***principles in machine learning***.
|
||||
The goal of this library is to push the extreme of the computation limits of machines to provide a ***scalable***, ***portable*** and ***accurate*** library.
|
||||
Make sure you [try it out](https://github.com/dmlc/xgboost), and most importantly, contribute your piece of wisdom (code, examples, tutorials) to the community!
|
||||
241
doc/parameter.md
241
doc/parameter.md
@ -1,241 +0,0 @@
|
||||
XGBoost Parameters
|
||||
==================
|
||||
Before running XGboost, we must set three types of parameters: general parameters, booster parameters and task parameters.
|
||||
- General parameters relates to which booster we are using to do boosting, commonly tree or linear model
|
||||
- Booster parameters depends on which booster you have chosen
|
||||
- Learning Task parameters that decides on the learning scenario, for example, regression tasks may use different parameters with ranking tasks.
|
||||
- Command line parameters that relates to behavior of CLI version of xgboost.
|
||||
|
||||
Parameters in R Package
|
||||
-----------------------
|
||||
In R-package, you can use .(dot) to replace underscore in the parameters, for example, you can use max.depth as max_depth. The underscore parameters are also valid in R.
|
||||
|
||||
General Parameters
|
||||
------------------
|
||||
* booster [default=gbtree]
|
||||
- which booster to use, can be gbtree, gblinear or dart. gbtree and dart use tree based model while gblinear uses linear function.
|
||||
* silent [default=0]
|
||||
- 0 means printing running messages, 1 means silent mode.
|
||||
* nthread [default to maximum number of threads available if not set]
|
||||
- number of parallel threads used to run xgboost
|
||||
* num_pbuffer [set automatically by xgboost, no need to be set by user]
|
||||
- size of prediction buffer, normally set to number of training instances. The buffers are used to save the prediction results of last boosting step.
|
||||
* num_feature [set automatically by xgboost, no need to be set by user]
|
||||
- feature dimension used in boosting, set to maximum dimension of the feature
|
||||
|
||||
Parameters for Tree Booster
|
||||
---------------------------
|
||||
* eta [default=0.3, alias: learning_rate]
|
||||
- step size shrinkage used in update to prevents overfitting. After each boosting step, we can directly get the weights of new features. and eta actually shrinks the feature weights to make the boosting process more conservative.
|
||||
- range: [0,1]
|
||||
* gamma [default=0, alias: min_split_loss]
|
||||
- minimum loss reduction required to make a further partition on a leaf node of the tree. The larger, the more conservative the algorithm will be.
|
||||
- range: [0,∞]
|
||||
* max_depth [default=6]
|
||||
- maximum depth of a tree, increase this value will make the model more complex / likely to be overfitting. 0 indicates no limit, limit is required for depth-wise grow policy.
|
||||
- range: [0,∞]
|
||||
* min_child_weight [default=1]
|
||||
- minimum sum of instance weight (hessian) needed in a child. If the tree partition step results in a leaf node with the sum of instance weight less than min_child_weight, then the building process will give up further partitioning. In linear regression mode, this simply corresponds to minimum number of instances needed to be in each node. The larger, the more conservative the algorithm will be.
|
||||
- range: [0,∞]
|
||||
* max_delta_step [default=0]
|
||||
- Maximum delta step we allow each tree's weight estimation to be. If the value is set to 0, it means there is no constraint. If it is set to a positive value, it can help making the update step more conservative. Usually this parameter is not needed, but it might help in logistic regression when class is extremely imbalanced. Set it to value of 1-10 might help control the update
|
||||
- range: [0,∞]
|
||||
* subsample [default=1]
|
||||
- subsample ratio of the training instance. Setting it to 0.5 means that XGBoost randomly collected half of the data instances to grow trees and this will prevent overfitting.
|
||||
- range: (0,1]
|
||||
* colsample_bytree [default=1]
|
||||
- subsample ratio of columns when constructing each tree.
|
||||
- range: (0,1]
|
||||
* colsample_bylevel [default=1]
|
||||
- subsample ratio of columns for each split, in each level.
|
||||
- range: (0,1]
|
||||
* lambda [default=1, alias: reg_lambda]
|
||||
- L2 regularization term on weights, increase this value will make model more conservative.
|
||||
* alpha [default=0, alias: reg_alpha]
|
||||
- L1 regularization term on weights, increase this value will make model more conservative.
|
||||
* tree_method, string [default='auto']
|
||||
- The tree construction algorithm used in XGBoost(see description in the [reference paper](http://arxiv.org/abs/1603.02754))
|
||||
- Distributed and external memory version only support approximate algorithm.
|
||||
- Choices: {'auto', 'exact', 'approx', 'hist', 'gpu_exact', 'gpu_hist'}
|
||||
- 'auto': Use heuristic to choose faster one.
|
||||
- For small to medium dataset, exact greedy will be used.
|
||||
- For very large-dataset, approximate algorithm will be chosen.
|
||||
- Because old behavior is always use exact greedy in single machine,
|
||||
user will get a message when approximate algorithm is chosen to notify this choice.
|
||||
- 'exact': Exact greedy algorithm.
|
||||
- 'approx': Approximate greedy algorithm using sketching and histogram.
|
||||
- 'hist': Fast histogram optimized approximate greedy algorithm. It uses some performance improvements such as bins caching.
|
||||
- 'gpu_exact': GPU implementation of exact algorithm.
|
||||
- 'gpu_hist': GPU implementation of hist algorithm.
|
||||
* sketch_eps, [default=0.03]
|
||||
- This is only used for approximate greedy algorithm.
|
||||
- This roughly translated into ```O(1 / sketch_eps)``` number of bins.
|
||||
Compared to directly select number of bins, this comes with theoretical guarantee with sketch accuracy.
|
||||
- Usually user does not have to tune this.
|
||||
but consider setting to a lower number for more accurate enumeration.
|
||||
- range: (0, 1)
|
||||
* scale_pos_weight, [default=1]
|
||||
- Control the balance of positive and negative weights, useful for unbalanced classes. A typical value to consider: sum(negative cases) / sum(positive cases) See [Parameters Tuning](how_to/param_tuning.md) for more discussion. Also see Higgs Kaggle competition demo for examples: [R](../demo/kaggle-higgs/higgs-train.R ), [py1](../demo/kaggle-higgs/higgs-numpy.py ), [py2](../demo/kaggle-higgs/higgs-cv.py ), [py3](../demo/guide-python/cross_validation.py)
|
||||
* updater, [default='grow_colmaker,prune']
|
||||
- A comma separated string defining the sequence of tree updaters to run, providing a modular way to construct and to modify the trees. This is an advanced parameter that is usually set automatically, depending on some other parameters. However, it could be also set explicitly by a user. The following updater plugins exist:
|
||||
- 'grow_colmaker': non-distributed column-based construction of trees.
|
||||
- 'distcol': distributed tree construction with column-based data splitting mode.
|
||||
- 'grow_histmaker': distributed tree construction with row-based data splitting based on global proposal of histogram counting.
|
||||
- 'grow_local_histmaker': based on local histogram counting.
|
||||
- 'grow_skmaker': uses the approximate sketching algorithm.
|
||||
- 'sync': synchronizes trees in all distributed nodes.
|
||||
- 'refresh': refreshes tree's statistics and/or leaf values based on the current data. Note that no random subsampling of data rows is performed.
|
||||
- 'prune': prunes the splits where loss < min_split_loss (or gamma).
|
||||
- In a distributed setting, the implicit updater sequence value would be adjusted as follows:
|
||||
- 'grow_histmaker,prune' when dsplit='row' (or default); or when data has multiple sparse pages
|
||||
- 'distcol' when dsplit='col'
|
||||
* refresh_leaf, [default=1]
|
||||
- This is a parameter of the 'refresh' updater plugin. When this flag is true, tree leafs as well as tree nodes' stats are updated. When it is false, only node stats are updated.
|
||||
* process_type, [default='default']
|
||||
- A type of boosting process to run.
|
||||
- Choices: {'default', 'update'}
|
||||
- 'default': the normal boosting process which creates new trees.
|
||||
- 'update': starts from an existing model and only updates its trees. In each boosting iteration, a tree from the initial model is taken, a specified sequence of updater plugins is run for that tree, and a modified tree is added to the new model. The new model would have either the same or smaller number of trees, depending on the number of boosting iteratons performed. Currently, the following built-in updater plugins could be meaningfully used with this process type: 'refresh', 'prune'. With 'update', one cannot use updater plugins that create new trees.
|
||||
* grow_policy, string [default='depthwise']
|
||||
- Controls a way new nodes are added to the tree.
|
||||
- Currently supported only if `tree_method` is set to 'hist'.
|
||||
- Choices: {'depthwise', 'lossguide'}
|
||||
- 'depthwise': split at nodes closest to the root.
|
||||
- 'lossguide': split at nodes with highest loss change.
|
||||
* max_leaves, [default=0]
|
||||
- Maximum number of nodes to be added. Only relevant for the 'lossguide' grow policy.
|
||||
* max_bin, [default=256]
|
||||
- This is only used if 'hist' is specified as `tree_method`.
|
||||
- Maximum number of discrete bins to bucket continuous features.
|
||||
- Increasing this number improves the optimality of splits at the cost of higher computation time.
|
||||
* predictor, [default='cpu_predictor']
|
||||
- The type of predictor algorithm to use. Provides the same results but allows the use of GPU or CPU.
|
||||
- 'cpu_predictor': Multicore CPU prediction algorithm.
|
||||
- 'gpu_predictor': Prediction using GPU. Default for 'gpu_exact' and 'gpu_hist' tree method.
|
||||
|
||||
Additional parameters for Dart Booster
|
||||
--------------------------------------
|
||||
* sample_type [default="uniform"]
|
||||
- type of sampling algorithm.
|
||||
- "uniform": dropped trees are selected uniformly.
|
||||
- "weighted": dropped trees are selected in proportion to weight.
|
||||
* normalize_type [default="tree"]
|
||||
- type of normalization algorithm.
|
||||
- "tree": new trees have the same weight of each of dropped trees.
|
||||
- weight of new trees are 1 / (k + learning_rate)
|
||||
- dropped trees are scaled by a factor of k / (k + learning_rate)
|
||||
- "forest": new trees have the same weight of sum of dropped trees (forest).
|
||||
- weight of new trees are 1 / (1 + learning_rate)
|
||||
- dropped trees are scaled by a factor of 1 / (1 + learning_rate)
|
||||
* rate_drop [default=0.0]
|
||||
- dropout rate (a fraction of previous trees to drop during the dropout).
|
||||
- range: [0.0, 1.0]
|
||||
* one_drop [default=0]
|
||||
- when this flag is enabled, at least one tree is always dropped during the dropout (allows Binomial-plus-one or epsilon-dropout from the original DART paper).
|
||||
* skip_drop [default=0.0]
|
||||
- Probability of skipping the dropout procedure during a boosting iteration.
|
||||
- If a dropout is skipped, new trees are added in the same manner as gbtree.
|
||||
- Note that non-zero skip_drop has higher priority than rate_drop or one_drop.
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
Parameters for Linear Booster
|
||||
-----------------------------
|
||||
* lambda [default=0, alias: reg_lambda]
|
||||
- L2 regularization term on weights, increase this value will make model more conservative. Normalised to number of training examples.
|
||||
* alpha [default=0, alias: reg_alpha]
|
||||
- L1 regularization term on weights, increase this value will make model more conservative. Normalised to number of training examples.
|
||||
* updater [default='shotgun']
|
||||
- Linear model algorithm
|
||||
- 'shotgun': Parallel coordinate descent algorithm based on shotgun algorithm. Uses 'hogwild' parallelism and therefore produces a nondeterministic solution on each run.
|
||||
- 'coord_descent': Ordinary coordinate descent algorithm. Also multithreaded but still produces a deterministic solution.
|
||||
|
||||
|
||||
Parameters for Tweedie Regression
|
||||
---------------------------------
|
||||
* tweedie_variance_power [default=1.5]
|
||||
- parameter that controls the variance of the Tweedie distribution
|
||||
- var(y) ~ E(y)^tweedie_variance_power
|
||||
- range: (1,2)
|
||||
- set closer to 2 to shift towards a gamma distribution
|
||||
- set closer to 1 to shift towards a Poisson distribution.
|
||||
|
||||
Learning Task Parameters
|
||||
------------------------
|
||||
Specify the learning task and the corresponding learning objective. The objective options are below:
|
||||
* objective [default=reg:linear]
|
||||
- "reg:linear" --linear regression
|
||||
- "reg:logistic" --logistic regression
|
||||
- "binary:logistic" --logistic regression for binary classification, output probability
|
||||
- "binary:logitraw" --logistic regression for binary classification, output score before logistic transformation
|
||||
- "gpu:reg:linear", "gpu:reg:logistic", "gpu:binary:logistic", gpu:binary:logitraw" --versions
|
||||
of the corresponding objective functions evaluated on the GPU; note that like the GPU histogram algorithm,
|
||||
they can only be used when the entire training session uses the same dataset
|
||||
- "count:poisson" --poisson regression for count data, output mean of poisson distribution
|
||||
- max_delta_step is set to 0.7 by default in poisson regression (used to safeguard optimization)
|
||||
- "survival:cox" --Cox regression for right censored survival time data (negative values are considered right censored).
|
||||
Note that predictions are returned on the hazard ratio scale (i.e., as HR = exp(marginal_prediction) in the proportional hazard function h(t) = h0(t) * HR).
|
||||
- "multi:softmax" --set XGBoost to do multiclass classification using the softmax objective, you also need to set num_class(number of classes)
|
||||
- "multi:softprob" --same as softmax, but output a vector of ndata * nclass, which can be further reshaped to ndata, nclass matrix. The result contains predicted probability of each data point belonging to each class.
|
||||
- "rank:pairwise" --set XGBoost to do ranking task by minimizing the pairwise loss
|
||||
- "reg:gamma" --gamma regression with log-link. Output is a mean of gamma distribution. It might be useful, e.g., for modeling insurance claims severity, or for any outcome that might be [gamma-distributed](https://en.wikipedia.org/wiki/Gamma_distribution#Applications)
|
||||
- "reg:tweedie" --Tweedie regression with log-link. It might be useful, e.g., for modeling total loss in insurance, or for any outcome that might be [Tweedie-distributed](https://en.wikipedia.org/wiki/Tweedie_distribution#Applications).
|
||||
* base_score [default=0.5]
|
||||
- the initial prediction score of all instances, global bias
|
||||
- for sufficient number of iterations, changing this value will not have too much effect.
|
||||
* eval_metric [default according to objective]
|
||||
- evaluation metrics for validation data, a default metric will be assigned according to objective (rmse for regression, and error for classification, mean average precision for ranking )
|
||||
- User can add multiple evaluation metrics, for python user, remember to pass the metrics in as list of parameters pairs instead of map, so that latter 'eval_metric' won't override previous one
|
||||
- The choices are listed below:
|
||||
- "rmse": [root mean square error](http://en.wikipedia.org/wiki/Root_mean_square_error)
|
||||
- "mae": [mean absolute error](https://en.wikipedia.org/wiki/Mean_absolute_error)
|
||||
- "logloss": negative [log-likelihood](http://en.wikipedia.org/wiki/Log-likelihood)
|
||||
- "error": Binary classification error rate. It is calculated as #(wrong cases)/#(all cases). For the predictions, the evaluation will regard the instances with prediction value larger than 0.5 as positive instances, and the others as negative instances.
|
||||
- "error@t": a different than 0.5 binary classification threshold value could be specified by providing a numerical value through 't'.
|
||||
- "merror": Multiclass classification error rate. It is calculated as #(wrong cases)/#(all cases).
|
||||
- "mlogloss": [Multiclass logloss](https://www.kaggle.com/wiki/LogLoss)
|
||||
- "auc": [Area under the curve](http://en.wikipedia.org/wiki/Receiver_operating_characteristic#Area_under_curve) for ranking evaluation.
|
||||
- "ndcg":[Normalized Discounted Cumulative Gain](http://en.wikipedia.org/wiki/NDCG)
|
||||
- "map":[Mean average precision](http://en.wikipedia.org/wiki/Mean_average_precision#Mean_average_precision)
|
||||
- "ndcg@n","map@n": n can be assigned as an integer to cut off the top positions in the lists for evaluation.
|
||||
- "ndcg-","map-","ndcg@n-","map@n-": In XGBoost, NDCG and MAP will evaluate the score of a list without any positive samples as 1. By adding "-" in the evaluation metric XGBoost will evaluate these score as 0 to be consistent under some conditions.
|
||||
training repeatedly
|
||||
- "poisson-nloglik": negative log-likelihood for Poisson regression
|
||||
- "gamma-nloglik": negative log-likelihood for gamma regression
|
||||
- "cox-nloglik": negative partial log-likelihood for Cox proportional hazards regression
|
||||
- "gamma-deviance": residual deviance for gamma regression
|
||||
- "tweedie-nloglik": negative log-likelihood for Tweedie regression (at a specified value of the tweedie_variance_power parameter)
|
||||
* seed [default=0]
|
||||
- random number seed.
|
||||
|
||||
Command Line Parameters
|
||||
-----------------------
|
||||
The following parameters are only used in the console version of xgboost
|
||||
* use_buffer [default=1]
|
||||
- Whether to create a binary buffer from text input. Doing so normally will speed up loading times
|
||||
* num_round
|
||||
- The number of rounds for boosting
|
||||
* data
|
||||
- The path of training data
|
||||
* test:data
|
||||
- The path of test data to do prediction
|
||||
* save_period [default=0]
|
||||
- the period to save the model, setting save_period=10 means that for every 10 rounds XGBoost will save the model, setting it to 0 means not saving any model during the training.
|
||||
* task [default=train] options: train, pred, eval, dump
|
||||
- train: training using data
|
||||
- pred: making prediction for test:data
|
||||
- eval: for evaluating statistics specified by eval[name]=filename
|
||||
- dump: for dump the learned model into text format (preliminary)
|
||||
* model_in [default=NULL]
|
||||
- path to input model, needed for test, eval, dump, if it is specified in training, xgboost will continue training from the input model
|
||||
* model_out [default=NULL]
|
||||
- path to output model after training finishes, if not specified, will output like 0003.model where 0003 is number of rounds to do boosting.
|
||||
* model_dir [default=models]
|
||||
- The output directory of the saved models during training
|
||||
* fmap
|
||||
- feature map, used for dump model
|
||||
* name_dump [default=dump.txt]
|
||||
- name of model dump file
|
||||
* name_pred [default=pred.txt]
|
||||
- name of prediction file, used in pred mode
|
||||
* pred_margin [default=0]
|
||||
- predict margin instead of transformed probability
|
||||
357
doc/parameter.rst
Normal file
357
doc/parameter.rst
Normal file
@ -0,0 +1,357 @@
|
||||
##################
|
||||
XGBoost Parameters
|
||||
##################
|
||||
Before running XGBoost, we must set three types of parameters: general parameters, booster parameters and task parameters.
|
||||
|
||||
- **General parameters** relate to which booster we are using to do boosting, commonly tree or linear model
|
||||
- **Booster parameters** depend on which booster you have chosen
|
||||
- **Learning task parameters** decide on the learning scenario. For example, regression tasks may use different parameters with ranking tasks.
|
||||
- **Command line parameters** relate to behavior of CLI version of XGBoost.
|
||||
|
||||
.. note:: Parameters in R package
|
||||
|
||||
In R-package, you can use ``.`` (dot) to replace underscore in the parameters, for example, you can use ``max.depth`` to indicate ``max_depth``. The underscore parameters are also valid in R.
|
||||
|
||||
******************
|
||||
General Parameters
|
||||
******************
|
||||
* ``booster`` [default= ``gbtree`` ]
|
||||
|
||||
- Which booster to use. Can be ``gbtree``, ``gblinear`` or ``dart``; ``gbtree`` and ``dart`` use tree based models while ``gblinear`` uses linear functions.
|
||||
|
||||
* ``silent`` [default=0]
|
||||
|
||||
- 0 means printing running messages, 1 means silent mode
|
||||
|
||||
* ``nthread`` [default to maximum number of threads available if not set]
|
||||
|
||||
- Number of parallel threads used to run XGBoost
|
||||
|
||||
* ``num_pbuffer`` [set automatically by XGBoost, no need to be set by user]
|
||||
|
||||
- Size of prediction buffer, normally set to number of training instances. The buffers are used to save the prediction results of last boosting step.
|
||||
|
||||
* ``num_feature`` [set automatically by XGBoost, no need to be set by user]
|
||||
|
||||
- Feature dimension used in boosting, set to maximum dimension of the feature
|
||||
|
||||
Parameters for Tree Booster
|
||||
===========================
|
||||
* ``eta`` [default=0.3, alias: ``learning_rate``]
|
||||
|
||||
- Step size shrinkage used in update to prevents overfitting. After each boosting step, we can directly get the weights of new features, and ``eta`` shrinks the feature weights to make the boosting process more conservative.
|
||||
- range: [0,1]
|
||||
|
||||
* ``gamma`` [default=0, alias: ``min_split_loss``]
|
||||
|
||||
- Minimum loss reduction required to make a further partition on a leaf node of the tree. The larger ``gamma`` is, the more conservative the algorithm will be.
|
||||
- range: [0,∞]
|
||||
|
||||
* ``max_depth`` [default=6]
|
||||
|
||||
- Maximum depth of a tree. Increasing this value will make the model more complex and more likely to overfit. 0 indicates no limit. Note that limit is required when ``grow_policy`` is set of ``depthwise``.
|
||||
- range: [0,∞]
|
||||
|
||||
* ``min_child_weight`` [default=1]
|
||||
|
||||
- Minimum sum of instance weight (hessian) needed in a child. If the tree partition step results in a leaf node with the sum of instance weight less than ``min_child_weight``, then the building process will give up further partitioning. In linear regression task, this simply corresponds to minimum number of instances needed to be in each node. The larger ``min_child_weight`` is, the more conservative the algorithm will be.
|
||||
- range: [0,∞]
|
||||
|
||||
* ``max_delta_step`` [default=0]
|
||||
|
||||
- Maximum delta step we allow each leaf output to be. If the value is set to 0, it means there is no constraint. If it is set to a positive value, it can help making the update step more conservative. Usually this parameter is not needed, but it might help in logistic regression when class is extremely imbalanced. Set it to value of 1-10 might help control the update.
|
||||
- range: [0,∞]
|
||||
|
||||
* ``subsample`` [default=1]
|
||||
|
||||
- Subsample ratio of the training instances. Setting it to 0.5 means that XGBoost would randomly sample half of the training data prior to growing trees. and this will prevent overfitting. Subsampling will occur once in every boosting iteration.
|
||||
- range: (0,1]
|
||||
|
||||
* ``colsample_bytree`` [default=1]
|
||||
|
||||
- Subsample ratio of columns when constructing each tree. Subsampling will occur once in every boosting iteration.
|
||||
- range: (0,1]
|
||||
|
||||
* ``colsample_bylevel`` [default=1]
|
||||
|
||||
- Subsample ratio of columns for each split, in each level. Subsampling will occur each time a new split is made. This paramter has no effect when ``tree_method`` is set to ``hist``.
|
||||
- range: (0,1]
|
||||
|
||||
* ``lambda`` [default=1, alias: ``reg_lambda``]
|
||||
|
||||
- L2 regularization term on weights. Increasing this value will make model more conservative.
|
||||
|
||||
* ``alpha`` [default=0, alias: ``reg_alpha``]
|
||||
|
||||
- L1 regularization term on weights. Increasing this value will make model more conservative.
|
||||
|
||||
* ``tree_method`` string [default= ``auto``]
|
||||
|
||||
- The tree construction algorithm used in XGBoost. See description in the `reference paper <http://arxiv.org/abs/1603.02754>`_.
|
||||
- Distributed and external memory version only support ``tree_method=approx``.
|
||||
- Choices: ``auto``, ``exact``, ``approx``, ``hist``, ``gpu_exact``, ``gpu_hist``
|
||||
|
||||
- ``auto``: Use heuristic to choose the fastest method.
|
||||
|
||||
- For small to medium dataset, exact greedy (``exact``) will be used.
|
||||
- For very large dataset, approximate algorithm (``approx``) will be chosen.
|
||||
- Because old behavior is always use exact greedy in single machine,
|
||||
user will get a message when approximate algorithm is chosen to notify this choice.
|
||||
|
||||
- ``exact``: Exact greedy algorithm.
|
||||
- ``approx``: Approximate greedy algorithm using quantile sketch and gradient histogram.
|
||||
- ``hist``: Fast histogram optimized approximate greedy algorithm. It uses some performance improvements such as bins caching.
|
||||
- ``gpu_exact``: GPU implementation of ``exact`` algorithm.
|
||||
- ``gpu_hist``: GPU implementation of ``hist`` algorithm.
|
||||
|
||||
* ``sketch_eps`` [default=0.03]
|
||||
|
||||
- Only used for ``tree_method=approx``.
|
||||
- This roughly translates into ``O(1 / sketch_eps)`` number of bins.
|
||||
Compared to directly select number of bins, this comes with theoretical guarantee with sketch accuracy.
|
||||
- Usually user does not have to tune this.
|
||||
But consider setting to a lower number for more accurate enumeration of split candidates.
|
||||
- range: (0, 1)
|
||||
|
||||
* ``scale_pos_weight`` [default=1]
|
||||
|
||||
- Control the balance of positive and negative weights, useful for unbalanced classes. A typical value to consider: ``sum(negative instances) / sum(positive instances)``. See `Parameters Tuning </tutorials/param_tuning>`_ for more discussion. Also, see Higgs Kaggle competition demo for examples: `R <https://github.com/dmlc/xgboost/blob/master/demo/kaggle-higgs/higgs-train.R>`_, `py1 <https://github.com/dmlc/xgboost/blob/master/demo/kaggle-higgs/higgs-numpy.py>`_, `py2 <https://github.com/dmlc/xgboost/blob/master/demo/kaggle-higgs/higgs-cv.py>`_, `py3 <https://github.com/dmlc/xgboost/blob/master/demo/guide-python/cross_validation.py>`_.
|
||||
|
||||
* ``updater`` [default= ``grow_colmaker,prune``]
|
||||
|
||||
- A comma separated string defining the sequence of tree updaters to run, providing a modular way to construct and to modify the trees. This is an advanced parameter that is usually set automatically, depending on some other parameters. However, it could be also set explicitly by a user. The following updater plugins exist:
|
||||
|
||||
- ``grow_colmaker``: non-distributed column-based construction of trees.
|
||||
- ``distcol``: distributed tree construction with column-based data splitting mode.
|
||||
- ``grow_histmaker``: distributed tree construction with row-based data splitting based on global proposal of histogram counting.
|
||||
- ``grow_local_histmaker``: based on local histogram counting.
|
||||
- ``grow_skmaker``: uses the approximate sketching algorithm.
|
||||
- ``sync``: synchronizes trees in all distributed nodes.
|
||||
- ``refresh``: refreshes tree's statistics and/or leaf values based on the current data. Note that no random subsampling of data rows is performed.
|
||||
- ``prune``: prunes the splits where loss < min_split_loss (or gamma).
|
||||
|
||||
- In a distributed setting, the implicit updater sequence value would be adjusted to ``grow_histmaker,prune``.
|
||||
|
||||
* ``refresh_leaf`` [default=1]
|
||||
|
||||
- This is a parameter of the ``refresh`` updater plugin. When this flag is 1, tree leafs as well as tree nodes' stats are updated. When it is 0, only node stats are updated.
|
||||
|
||||
* ``process_type`` [default= ``default``]
|
||||
|
||||
- A type of boosting process to run.
|
||||
- Choices: ``default``, ``update``
|
||||
|
||||
- ``default``: The normal boosting process which creates new trees.
|
||||
- ``update``: Starts from an existing model and only updates its trees. In each boosting iteration, a tree from the initial model is taken, a specified sequence of updater plugins is run for that tree, and a modified tree is added to the new model. The new model would have either the same or smaller number of trees, depending on the number of boosting iteratons performed. Currently, the following built-in updater plugins could be meaningfully used with this process type: ``refresh``, ``prune``. With ``process_type=update``, one cannot use updater plugins that create new trees.
|
||||
|
||||
* ``grow_policy`` [default= ``depthwise``]
|
||||
|
||||
- Controls a way new nodes are added to the tree.
|
||||
- Currently supported only if ``tree_method`` is set to ``hist``.
|
||||
- Choices: ``depthwise``, ```lossguide``
|
||||
|
||||
- ``depthwise``: split at nodes closest to the root.
|
||||
- ``lossguide``: split at nodes with highest loss change.
|
||||
|
||||
* ``max_leaves`` [default=0]
|
||||
|
||||
- Maximum number of nodes to be added. Only relevant when ``grow_policy=lossguide`` is set.
|
||||
|
||||
* ``max_bin``, [default=256]
|
||||
|
||||
- Only used if ``tree_method`` is set to ``hist``.
|
||||
- Maximum number of discrete bins to bucket continuous features.
|
||||
- Increasing this number improves the optimality of splits at the cost of higher computation time.
|
||||
|
||||
* ``predictor``, [default=``cpu_predictor``]
|
||||
|
||||
- The type of predictor algorithm to use. Provides the same results but allows the use of GPU or CPU.
|
||||
|
||||
- ``cpu_predictor``: Multicore CPU prediction algorithm.
|
||||
- ``gpu_predictor``: Prediction using GPU. Default when ``tree_method`` is ``gpu_exact`` or ``gpu_hist``.
|
||||
|
||||
Additional parameters for Dart Booster (``booster=dart``)
|
||||
=========================================================
|
||||
* ``sample_type`` [default= ``uniform``]
|
||||
|
||||
- Type of sampling algorithm.
|
||||
|
||||
- ``uniform``: dropped trees are selected uniformly.
|
||||
- ``weighted``: dropped trees are selected in proportion to weight.
|
||||
|
||||
* ``normalize_type`` [default= ``tree``]
|
||||
|
||||
- Type of normalization algorithm.
|
||||
|
||||
- ``tree``: new trees have the same weight of each of dropped trees.
|
||||
|
||||
- Weight of new trees are ``1 / (k + learning_rate)``.
|
||||
- Dropped trees are scaled by a factor of ``k / (k + learning_rate)``.
|
||||
|
||||
- ``forest``: new trees have the same weight of sum of dropped trees (forest).
|
||||
|
||||
- Weight of new trees are ``1 / (1 + learning_rate)``.
|
||||
- Dropped trees are scaled by a factor of ``1 / (1 + learning_rate)``.
|
||||
|
||||
* ``rate_drop`` [default=0.0]
|
||||
|
||||
- Dropout rate (a fraction of previous trees to drop during the dropout).
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
* ``one_drop`` [default=0]
|
||||
|
||||
- When this flag is enabled, at least one tree is always dropped during the dropout (allows Binomial-plus-one or epsilon-dropout from the original DART paper).
|
||||
|
||||
* ``skip_drop`` [default=0.0]
|
||||
|
||||
- Probability of skipping the dropout procedure during a boosting iteration.
|
||||
|
||||
- If a dropout is skipped, new trees are added in the same manner as ``gbtree``.
|
||||
- Note that non-zero ``skip_drop`` has higher priority than ``rate_drop`` or ``one_drop``.
|
||||
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
Parameters for Linear Booster (``booster=gbtree``)
|
||||
==================================================
|
||||
* ``lambda`` [default=0, alias: ``reg_lambda``]
|
||||
|
||||
- L2 regularization term on weights. Increasing this value will make model more conservative. Normalised to number of training examples.
|
||||
|
||||
* ``alpha`` [default=0, alias: ``reg_alpha``]
|
||||
|
||||
- L1 regularization term on weights. Increasing this value will make model more conservative. Normalised to number of training examples.
|
||||
|
||||
* ``updater`` [default= ``shotgun``]
|
||||
|
||||
- Choice of algorithm to fit linear model
|
||||
|
||||
- ``shotgun``: Parallel coordinate descent algorithm based on shotgun algorithm. Uses 'hogwild' parallelism and therefore produces a nondeterministic solution on each run.
|
||||
- ``coord_descent``: Ordinary coordinate descent algorithm. Also multithreaded but still produces a deterministic solution.
|
||||
|
||||
Parameters for Tweedie Regression (``objective=reg:tweedie``)
|
||||
=============================================================
|
||||
* ``tweedie_variance_power`` [default=1.5]
|
||||
|
||||
- Parameter that controls the variance of the Tweedie distribution ``var(y) ~ E(y)^tweedie_variance_power``
|
||||
- range: (1,2)
|
||||
- Set closer to 2 to shift towards a gamma distribution
|
||||
- Set closer to 1 to shift towards a Poisson distribution.
|
||||
|
||||
************************
|
||||
Learning Task Parameters
|
||||
************************
|
||||
Specify the learning task and the corresponding learning objective. The objective options are below:
|
||||
|
||||
* ``objective`` [default=reg:linear]
|
||||
|
||||
- ``reg:linear``: linear regression
|
||||
- ``reg:logistic``: logistic regression
|
||||
- ``binary:logistic``: logistic regression for binary classification, output probability
|
||||
- ``binary:logitraw``: logistic regression for binary classification, output score before logistic transformation
|
||||
- ``gpu:reg:linear``, ``gpu:reg:logistic``, ``gpu:binary:logistic``, ``gpu:binary:logitraw``: versions
|
||||
of the corresponding objective functions evaluated on the GPU; note that like the GPU histogram algorithm,
|
||||
they can only be used when the entire training session uses the same dataset
|
||||
- ``count:poisson`` --poisson regression for count data, output mean of poisson distribution
|
||||
|
||||
- ``max_delta_step`` is set to 0.7 by default in poisson regression (used to safeguard optimization)
|
||||
|
||||
- ``survival:cox``: Cox regression for right censored survival time data (negative values are considered right censored).
|
||||
Note that predictions are returned on the hazard ratio scale (i.e., as HR = exp(marginal_prediction) in the proportional hazard function ``h(t) = h0(t) * HR``).
|
||||
- ``multi:softmax``: set XGBoost to do multiclass classification using the softmax objective, you also need to set num_class(number of classes)
|
||||
- ``multi:softprob``: same as softmax, but output a vector of ``ndata * nclass``, which can be further reshaped to ``ndata * nclass`` matrix. The result contains predicted probability of each data point belonging to each class.
|
||||
- ``rank:pairwise``: set XGBoost to do ranking task by minimizing the pairwise loss
|
||||
- ``reg:gamma``: gamma regression with log-link. Output is a mean of gamma distribution. It might be useful, e.g., for modeling insurance claims severity, or for any outcome that might be `gamma-distributed <https://en.wikipedia.org/wiki/Gamma_distribution#Applications>`_.
|
||||
- ``reg:tweedie``: Tweedie regression with log-link. It might be useful, e.g., for modeling total loss in insurance, or for any outcome that might be `Tweedie-distributed <https://en.wikipedia.org/wiki/Tweedie_distribution#Applications>`_.
|
||||
|
||||
* ``base_score`` [default=0.5]
|
||||
|
||||
- The initial prediction score of all instances, global bias
|
||||
- For sufficient number of iterations, changing this value will not have too much effect.
|
||||
|
||||
* ``eval_metric`` [default according to objective]
|
||||
|
||||
- Evaluation metrics for validation data, a default metric will be assigned according to objective (rmse for regression, and error for classification, mean average precision for ranking)
|
||||
- User can add multiple evaluation metrics. Python users: remember to pass the metrics in as list of parameters pairs instead of map, so that latter ``eval_metric`` won't override previous one
|
||||
- The choices are listed below:
|
||||
|
||||
- ``rmse``: `root mean square error <http://en.wikipedia.org/wiki/Root_mean_square_error>`_
|
||||
- ``mae``: `mean absolute error <https://en.wikipedia.org/wiki/Mean_absolute_error>`_
|
||||
- ``logloss``: `negative log-likelihood <http://en.wikipedia.org/wiki/Log-likelihood>`_
|
||||
- ``error``: Binary classification error rate. It is calculated as ``#(wrong cases)/#(all cases)``. For the predictions, the evaluation will regard the instances with prediction value larger than 0.5 as positive instances, and the others as negative instances.
|
||||
- ``error@t``: a different than 0.5 binary classification threshold value could be specified by providing a numerical value through 't'.
|
||||
- ``merror``: Multiclass classification error rate. It is calculated as ``#(wrong cases)/#(all cases)``.
|
||||
- ``mlogloss``: `Multiclass logloss <https://www.kaggle.com/wiki/LogLoss>`_.
|
||||
- ``auc``: `Area under the curve <http://en.wikipedia.org/wiki/Receiver_operating_characteristic#Area_under_curve>`_
|
||||
- ``ndcg``: `Normalized Discounted Cumulative Gain <http://en.wikipedia.org/wiki/NDCG>`_
|
||||
- ``map``: `Mean average precision <http://en.wikipedia.org/wiki/Mean_average_precision#Mean_average_precision>`_
|
||||
- ``ndcg@n``, ``map@n``: 'n' can be assigned as an integer to cut off the top positions in the lists for evaluation.
|
||||
- ``ndcg-``, ``map-``, ``ndcg@n-``, ``map@n-``: In XGBoost, NDCG and MAP will evaluate the score of a list without any positive samples as 1. By adding "-" in the evaluation metric XGBoost will evaluate these score as 0 to be consistent under some conditions.
|
||||
- ``poisson-nloglik``: negative log-likelihood for Poisson regression
|
||||
- ``gamma-nloglik``: negative log-likelihood for gamma regression
|
||||
- ``cox-nloglik``: negative partial log-likelihood for Cox proportional hazards regression
|
||||
- ``gamma-deviance``: residual deviance for gamma regression
|
||||
- ``tweedie-nloglik``: negative log-likelihood for Tweedie regression (at a specified value of the ``tweedie_variance_power`` parameter)
|
||||
|
||||
* ``seed`` [default=0]
|
||||
|
||||
- Random number seed.
|
||||
|
||||
***********************
|
||||
Command Line Parameters
|
||||
***********************
|
||||
The following parameters are only used in the console version of XGBoost
|
||||
|
||||
* ``use_buffer`` [default=1]
|
||||
|
||||
- Whether to create a binary buffer from text input. Doing so normally will speed up loading times
|
||||
|
||||
* ``num_round``
|
||||
|
||||
- The number of rounds for boosting
|
||||
|
||||
* ``data``
|
||||
|
||||
- The path of training data
|
||||
|
||||
* ``test:data``
|
||||
|
||||
- The path of test data to do prediction
|
||||
|
||||
* ``save_period`` [default=0]
|
||||
|
||||
- The period to save the model. Setting ``save_period=10`` means that for every 10 rounds XGBoost will save the model. Setting it to 0 means not saving any model during the training.
|
||||
|
||||
* ``task`` [default= ``train``] options: ``train``, ``pred``, ``eval``, ``dump``
|
||||
|
||||
- ``train``: training using data
|
||||
- ``pred``: making prediction for test:data
|
||||
- ``eval``: for evaluating statistics specified by ``eval[name]=filename``
|
||||
- ``dump``: for dump the learned model into text format
|
||||
|
||||
* ``model_in`` [default=NULL]
|
||||
|
||||
- Path to input model, needed for ``test``, ``eval``, ``dump`` tasks. If it is specified in training, XGBoost will continue training from the input model.
|
||||
|
||||
* ``model_out`` [default=NULL]
|
||||
|
||||
- Path to output model after training finishes. If not specified, XGBoost will output files with such names as ``0003.model`` where ``0003`` is number of boosting rounds.
|
||||
|
||||
* ``model_dir`` [default= ``models/``]
|
||||
|
||||
- The output directory of the saved models during training
|
||||
|
||||
* ``fmap``
|
||||
|
||||
- Feature map, used for dumping model
|
||||
|
||||
* ``name_dump`` [default= ``dump.txt``]
|
||||
|
||||
- Name of model dump file
|
||||
|
||||
* ``name_pred`` [default= ``pred.txt``]
|
||||
|
||||
- Name of prediction file, used in pred mode
|
||||
|
||||
* ``pred_margin`` [default=0]
|
||||
|
||||
- Predict margin instead of transformed probability
|
||||
@ -1,10 +0,0 @@
|
||||
XGBoost Python Package
|
||||
======================
|
||||
This page contains links to all the python related documents on python package.
|
||||
To install the package package, checkout [Build and Installation Instruction](../build.md).
|
||||
|
||||
Contents
|
||||
--------
|
||||
* [Python Overview Tutorial](python_intro.md)
|
||||
* [Learning to use XGBoost by Example](../../demo)
|
||||
* [Python API Reference](python_api.rst)
|
||||
14
doc/python/index.rst
Normal file
14
doc/python/index.rst
Normal file
@ -0,0 +1,14 @@
|
||||
######################
|
||||
XGBoost Python Package
|
||||
######################
|
||||
This page contains links to all the python related documents on python package.
|
||||
To install the package package, checkout :doc:`Installation Guide </build>`.
|
||||
|
||||
********
|
||||
Contents
|
||||
********
|
||||
|
||||
.. toctree::
|
||||
python_intro
|
||||
python_api
|
||||
Python examples <https://github.com/dmlc/xgboost/tree/master/demo/guide-python>
|
||||
@ -2,8 +2,6 @@ Python API Reference
|
||||
====================
|
||||
This page gives the Python API reference of xgboost, please also refer to Python Package Introduction for more information about python package.
|
||||
|
||||
The document in this page is automatically generated by sphinx. The content do not render at github, you can view it at http://xgboost.readthedocs.org/en/latest/python/python_api.html
|
||||
|
||||
Core Data Structure
|
||||
-------------------
|
||||
.. automodule:: xgboost.core
|
||||
|
||||
@ -1,173 +0,0 @@
|
||||
Python Package Introduction
|
||||
===========================
|
||||
This document gives a basic walkthrough of xgboost python package.
|
||||
|
||||
***List of other Helpful Links***
|
||||
* [Python walkthrough code collections](https://github.com/tqchen/xgboost/blob/master/demo/guide-python)
|
||||
* [Python API Reference](python_api.rst)
|
||||
|
||||
Install XGBoost
|
||||
---------------
|
||||
To install XGBoost, do the following:
|
||||
|
||||
* Run `make` in the root directory of the project
|
||||
* In the `python-package` directory, run
|
||||
```shell
|
||||
python setup.py install
|
||||
```
|
||||
|
||||
To verify your installation, try to `import xgboost` in Python.
|
||||
```python
|
||||
import xgboost as xgb
|
||||
```
|
||||
|
||||
Data Interface
|
||||
--------------
|
||||
The XGBoost python module is able to load data from:
|
||||
- libsvm txt format file
|
||||
- comma-separated values (CSV) file
|
||||
- Numpy 2D array
|
||||
- Scipy 2D sparse array, and
|
||||
- xgboost binary buffer file.
|
||||
|
||||
The data is stored in a ```DMatrix``` object.
|
||||
|
||||
* To load a libsvm text file or a XGBoost binary file into ```DMatrix```:
|
||||
```python
|
||||
dtrain = xgb.DMatrix('train.svm.txt')
|
||||
dtest = xgb.DMatrix('test.svm.buffer')
|
||||
```
|
||||
* To load a CSV file into ```DMatrix```:
|
||||
```python
|
||||
# label_column specifies the index of the column containing the true label
|
||||
dtrain = xgb.DMatrix('train.csv?format=csv&label_column=0')
|
||||
dtest = xgb.DMatrix('test.csv?format=csv&label_column=0')
|
||||
```
|
||||
(Note that XGBoost does not support categorical features; if your data contains
|
||||
categorical features, load it as a numpy array first and then perform
|
||||
[one-hot encoding](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OneHotEncoder.html).)
|
||||
|
||||
* To load a numpy array into ```DMatrix```:
|
||||
```python
|
||||
data = np.random.rand(5, 10) # 5 entities, each contains 10 features
|
||||
label = np.random.randint(2, size=5) # binary target
|
||||
dtrain = xgb.DMatrix(data, label=label)
|
||||
```
|
||||
* To load a scpiy.sparse array into ```DMatrix```:
|
||||
```python
|
||||
csr = scipy.sparse.csr_matrix((dat, (row, col)))
|
||||
dtrain = xgb.DMatrix(csr)
|
||||
```
|
||||
* Saving ```DMatrix``` into a XGBoost binary file will make loading faster:
|
||||
```python
|
||||
dtrain = xgb.DMatrix('train.svm.txt')
|
||||
dtrain.save_binary('train.buffer')
|
||||
```
|
||||
* Missing values can be replaced by a default value in the ```DMatrix``` constructor:
|
||||
```python
|
||||
dtrain = xgb.DMatrix(data, label=label, missing=-999.0)
|
||||
```
|
||||
* Weights can be set when needed:
|
||||
```python
|
||||
w = np.random.rand(5, 1)
|
||||
dtrain = xgb.DMatrix(data, label=label, missing=-999.0, weight=w)
|
||||
```
|
||||
|
||||
Setting Parameters
|
||||
------------------
|
||||
XGBoost can use either a list of pairs or a dictionary to set [parameters](../parameter.md). For instance:
|
||||
* Booster parameters
|
||||
```python
|
||||
param = {'max_depth': 2, 'eta': 1, 'silent': 1, 'objective': 'binary:logistic'}
|
||||
param['nthread'] = 4
|
||||
param['eval_metric'] = 'auc'
|
||||
```
|
||||
* You can also specify multiple eval metrics:
|
||||
```python
|
||||
param['eval_metric'] = ['auc', 'ams@0']
|
||||
|
||||
# alternatively:
|
||||
# plst = param.items()
|
||||
# plst += [('eval_metric', 'ams@0')]
|
||||
```
|
||||
|
||||
* Specify validations set to watch performance
|
||||
```python
|
||||
evallist = [(dtest, 'eval'), (dtrain, 'train')]
|
||||
```
|
||||
|
||||
Training
|
||||
--------
|
||||
|
||||
Training a model requires a parameter list and data set.
|
||||
```python
|
||||
num_round = 10
|
||||
bst = xgb.train(plst, dtrain, num_round, evallist)
|
||||
```
|
||||
After training, the model can be saved.
|
||||
```python
|
||||
bst.save_model('0001.model')
|
||||
```
|
||||
The model and its feature map can also be dumped to a text file.
|
||||
```python
|
||||
# dump model
|
||||
bst.dump_model('dump.raw.txt')
|
||||
# dump model with feature map
|
||||
bst.dump_model('dump.raw.txt', 'featmap.txt')
|
||||
```
|
||||
A saved model can be loaded as follows:
|
||||
```python
|
||||
bst = xgb.Booster({'nthread': 4}) # init model
|
||||
bst.load_model('model.bin') # load data
|
||||
```
|
||||
|
||||
Early Stopping
|
||||
--------------
|
||||
If you have a validation set, you can use early stopping to find the optimal number of boosting rounds.
|
||||
Early stopping requires at least one set in `evals`. If there's more than one, it will use the last.
|
||||
|
||||
`train(..., evals=evals, early_stopping_rounds=10)`
|
||||
|
||||
The model will train until the validation score stops improving. Validation error needs to decrease at least every `early_stopping_rounds` to continue training.
|
||||
|
||||
If early stopping occurs, the model will have three additional fields: `bst.best_score`, `bst.best_iteration` and `bst.best_ntree_limit`. Note that `train()` will return a model from the last iteration, not the best one.
|
||||
|
||||
This works with both metrics to minimize (RMSE, log loss, etc.) and to maximize (MAP, NDCG, AUC). Note that if you specify more than one evaluation metric the last one in `param['eval_metric']` is used for early stopping.
|
||||
|
||||
Prediction
|
||||
----------
|
||||
A model that has been trained or loaded can perform predictions on data sets.
|
||||
```python
|
||||
# 7 entities, each contains 10 features
|
||||
data = np.random.rand(7, 10)
|
||||
dtest = xgb.DMatrix(data)
|
||||
ypred = bst.predict(dtest)
|
||||
```
|
||||
|
||||
If early stopping is enabled during training, you can get predictions from the best iteration with `bst.best_ntree_limit`:
|
||||
```python
|
||||
ypred = bst.predict(dtest, ntree_limit=bst.best_ntree_limit)
|
||||
```
|
||||
|
||||
Plotting
|
||||
--------
|
||||
|
||||
You can use plotting module to plot importance and output tree.
|
||||
|
||||
To plot importance, use ``plot_importance``. This function requires ``matplotlib`` to be installed.
|
||||
|
||||
```python
|
||||
xgb.plot_importance(bst)
|
||||
```
|
||||
|
||||
To plot the output tree via ``matplotlib``, use ``plot_tree``, specifying the ordinal number of the target tree. This function requires ``graphviz`` and ``matplotlib``.
|
||||
|
||||
```python
|
||||
xgb.plot_tree(bst, num_trees=2)
|
||||
```
|
||||
|
||||
When you use ``IPython``, you can use the ``to_graphviz`` function, which converts the target tree to a ``graphviz`` instance. The ``graphviz`` instance is automatically rendered in ``IPython``.
|
||||
|
||||
```python
|
||||
xgb.to_graphviz(bst, num_trees=2)
|
||||
```
|
||||
203
doc/python/python_intro.rst
Normal file
203
doc/python/python_intro.rst
Normal file
@ -0,0 +1,203 @@
|
||||
###########################
|
||||
Python Package Introduction
|
||||
###########################
|
||||
This document gives a basic walkthrough of xgboost python package.
|
||||
|
||||
**List of other Helpful Links**
|
||||
|
||||
* `Python walkthrough code collections <https://github.com/tqchen/xgboost/blob/master/demo/guide-python>`_
|
||||
* :doc:`Python API Reference <python_api>`
|
||||
|
||||
Install XGBoost
|
||||
---------------
|
||||
To install XGBoost, follow instructions in :doc:`/build`.
|
||||
|
||||
To verify your installation, run the following in Python:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
import xgboost as xgb
|
||||
|
||||
Data Interface
|
||||
--------------
|
||||
The XGBoost python module is able to load data from:
|
||||
|
||||
- LibSVM text format file
|
||||
- Comma-separated values (CSV) file
|
||||
- NumPy 2D array
|
||||
- SciPy 2D sparse array, and
|
||||
- XGBoost binary buffer file.
|
||||
|
||||
(See :doc:`/tutorials/input_format` for detailed description of text input format.)
|
||||
|
||||
The data is stored in a :py:class:`DMatrix <xgboost.DMatrix>` object.
|
||||
|
||||
* To load a libsvm text file or a XGBoost binary file into :py:class:`DMatrix <xgboost.DMatrix>`:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
dtrain = xgb.DMatrix('train.svm.txt')
|
||||
dtest = xgb.DMatrix('test.svm.buffer')
|
||||
|
||||
* To load a CSV file into :py:class:`DMatrix <xgboost.DMatrix>`:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
# label_column specifies the index of the column containing the true label
|
||||
dtrain = xgb.DMatrix('train.csv?format=csv&label_column=0')
|
||||
dtest = xgb.DMatrix('test.csv?format=csv&label_column=0')
|
||||
|
||||
(Note that XGBoost does not support categorical features; if your data contains
|
||||
categorical features, load it as a NumPy array first and then perform
|
||||
`one-hot encoding <http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OneHotEncoder.html>`_.)
|
||||
|
||||
* To load a NumPy array into :py:class:`DMatrix <xgboost.DMatrix>`:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
data = np.random.rand(5, 10) # 5 entities, each contains 10 features
|
||||
label = np.random.randint(2, size=5) # binary target
|
||||
dtrain = xgb.DMatrix(data, label=label)
|
||||
|
||||
* To load a :py:mod:`scipy.sparse` array into :py:class:`DMatrix <xgboost.DMatrix>`:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
csr = scipy.sparse.csr_matrix((dat, (row, col)))
|
||||
dtrain = xgb.DMatrix(csr)
|
||||
|
||||
* Saving :py:class:`DMatrix <xgboost.DMatrix>` into a XGBoost binary file will make loading faster:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
dtrain = xgb.DMatrix('train.svm.txt')
|
||||
dtrain.save_binary('train.buffer')
|
||||
|
||||
* Missing values can be replaced by a default value in the :py:class:`DMatrix <xgboost.DMatrix>` constructor:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
dtrain = xgb.DMatrix(data, label=label, missing=-999.0)
|
||||
|
||||
* Weights can be set when needed:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
w = np.random.rand(5, 1)
|
||||
dtrain = xgb.DMatrix(data, label=label, missing=-999.0, weight=w)
|
||||
|
||||
Setting Parameters
|
||||
------------------
|
||||
XGBoost can use either a list of pairs or a dictionary to set :doc:`parameters </parameter>`. For instance:
|
||||
|
||||
* Booster parameters
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
param = {'max_depth': 2, 'eta': 1, 'silent': 1, 'objective': 'binary:logistic'}
|
||||
param['nthread'] = 4
|
||||
param['eval_metric'] = 'auc'
|
||||
|
||||
* You can also specify multiple eval metrics:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
param['eval_metric'] = ['auc', 'ams@0']
|
||||
|
||||
# alternatively:
|
||||
# plst = param.items()
|
||||
# plst += [('eval_metric', 'ams@0')]
|
||||
|
||||
* Specify validations set to watch performance
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
evallist = [(dtest, 'eval'), (dtrain, 'train')]
|
||||
|
||||
Training
|
||||
--------
|
||||
|
||||
Training a model requires a parameter list and data set.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
num_round = 10
|
||||
bst = xgb.train(param, dtrain, num_round, evallist)
|
||||
|
||||
After training, the model can be saved.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
bst.save_model('0001.model')
|
||||
|
||||
The model and its feature map can also be dumped to a text file.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
# dump model
|
||||
bst.dump_model('dump.raw.txt')
|
||||
# dump model with feature map
|
||||
bst.dump_model('dump.raw.txt', 'featmap.txt')
|
||||
|
||||
A saved model can be loaded as follows:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
bst = xgb.Booster({'nthread': 4}) # init model
|
||||
bst.load_model('model.bin') # load data
|
||||
|
||||
Early Stopping
|
||||
--------------
|
||||
If you have a validation set, you can use early stopping to find the optimal number of boosting rounds.
|
||||
Early stopping requires at least one set in ``evals``. If there's more than one, it will use the last.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
train(..., evals=evals, early_stopping_rounds=10)
|
||||
|
||||
The model will train until the validation score stops improving. Validation error needs to decrease at least every ``early_stopping_rounds`` to continue training.
|
||||
|
||||
If early stopping occurs, the model will have three additional fields: ``bst.best_score``, ``bst.best_iteration`` and ``bst.best_ntree_limit``. Note that :py:meth:`xgboost.train` will return a model from the last iteration, not the best one.
|
||||
|
||||
This works with both metrics to minimize (RMSE, log loss, etc.) and to maximize (MAP, NDCG, AUC). Note that if you specify more than one evaluation metric the last one in ``param['eval_metric']`` is used for early stopping.
|
||||
|
||||
Prediction
|
||||
----------
|
||||
A model that has been trained or loaded can perform predictions on data sets.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
# 7 entities, each contains 10 features
|
||||
data = np.random.rand(7, 10)
|
||||
dtest = xgb.DMatrix(data)
|
||||
ypred = bst.predict(dtest)
|
||||
|
||||
If early stopping is enabled during training, you can get predictions from the best iteration with ``bst.best_ntree_limit``:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
ypred = bst.predict(dtest, ntree_limit=bst.best_ntree_limit)
|
||||
|
||||
Plotting
|
||||
--------
|
||||
|
||||
You can use plotting module to plot importance and output tree.
|
||||
|
||||
To plot importance, use :py:meth:`xgboost.plot_importance`. This function requires ``matplotlib`` to be installed.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
xgb.plot_importance(bst)
|
||||
|
||||
To plot the output tree via ``matplotlib``, use :py:meth:`xgboost.plot_tree`, specifying the ordinal number of the target tree. This function requires ``graphviz`` and ``matplotlib``.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
xgb.plot_tree(bst, num_trees=2)
|
||||
|
||||
When you use ``IPython``, you can use the :py:meth:`xgboost.to_graphviz` function, which converts the target tree to a ``graphviz`` instance. The ``graphviz`` instance is automatically rendered in ``IPython``.
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
xgb.to_graphviz(bst, num_trees=2)
|
||||
|
||||
@ -1,3 +1,4 @@
|
||||
sphinx==1.5.6
|
||||
commonmark==0.5.4
|
||||
sphinx
|
||||
mock
|
||||
guzzle_sphinx_theme
|
||||
breathe
|
||||
|
||||
@ -7,24 +7,10 @@ import subprocess
|
||||
|
||||
READTHEDOCS_BUILD = (os.environ.get('READTHEDOCS', None) is not None)
|
||||
|
||||
if not os.path.exists('../recommonmark'):
|
||||
subprocess.call('cd ..; rm -rf recommonmark;' +
|
||||
'git clone https://github.com/tqchen/recommonmark', shell = True)
|
||||
else:
|
||||
subprocess.call('cd ../recommonmark/; git pull', shell=True)
|
||||
|
||||
if not os.path.exists('web-data'):
|
||||
subprocess.call('rm -rf web-data;' +
|
||||
'git clone https://github.com/dmlc/web-data', shell = True)
|
||||
subprocess.call('rm -rf web-data;' +
|
||||
'git clone https://github.com/dmlc/web-data', shell = True)
|
||||
else:
|
||||
subprocess.call('cd web-data; git pull', shell=True)
|
||||
subprocess.call('cd web-data; git pull', shell=True)
|
||||
|
||||
|
||||
sys.path.insert(0, os.path.abspath('../recommonmark/'))
|
||||
sys.stderr.write('READTHEDOCS=%s\n' % (READTHEDOCS_BUILD))
|
||||
|
||||
|
||||
from recommonmark import parser, transform
|
||||
|
||||
MarkdownParser = parser.CommonMarkParser
|
||||
AutoStructify = transform.AutoStructify
|
||||
|
||||
@ -1,187 +0,0 @@
|
||||
Distributed XGBoost YARN on AWS
|
||||
===============================
|
||||
This is a step-by-step tutorial on how to setup and run distributed [XGBoost](https://github.com/dmlc/xgboost)
|
||||
on an AWS EC2 cluster. Distributed XGBoost runs on various platforms such as MPI, SGE and Hadoop YARN.
|
||||
In this tutorial, we use YARN as an example since this is a widely used solution for distributed computing.
|
||||
|
||||
Prerequisite
|
||||
------------
|
||||
We need to get a [AWS key-pair](http://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ec2-key-pairs.html)
|
||||
to access the AWS services. Let us assume that we are using a key ```mykey``` and the corresponding permission file ```mypem.pem```.
|
||||
|
||||
We also need [AWS credentials](http://docs.aws.amazon.com/AWSSimpleQueueService/latest/SQSGettingStartedGuide/AWSCredentials.html),
|
||||
which includes an `ACCESS_KEY_ID` and a `SECRET_ACCESS_KEY`.
|
||||
|
||||
Finally, we will need a S3 bucket to host the data and the model, ```s3://mybucket/```
|
||||
|
||||
Setup a Hadoop YARN Cluster
|
||||
---------------------------
|
||||
This sections shows how to start a Hadoop YARN cluster from scratch.
|
||||
You can skip this step if you have already have one.
|
||||
We will be using [yarn-ec2](https://github.com/tqchen/yarn-ec2) to start the cluster.
|
||||
|
||||
We can first clone the yarn-ec2 script by the following command.
|
||||
```bash
|
||||
git clone https://github.com/tqchen/yarn-ec2
|
||||
```
|
||||
|
||||
To use the script, we must set the environment variables `AWS_ACCESS_KEY_ID` and
|
||||
`AWS_SECRET_ACCESS_KEY` properly. This can be done by adding the following two lines in
|
||||
`~/.bashrc` (replacing the strings with the correct ones)
|
||||
|
||||
```bash
|
||||
export AWS_ACCESS_KEY_ID=AKIAIOSFODNN7EXAMPLE
|
||||
export AWS_SECRET_ACCESS_KEY=wJalrXUtnFEMI/K7MDENG/bPxRfiCYEXAMPLEKEY
|
||||
```
|
||||
|
||||
Now we can launch a master machine of the cluster from EC2
|
||||
```bash
|
||||
./yarn-ec2 -k mykey -i mypem.pem launch xgboost
|
||||
```
|
||||
Wait a few mininutes till the master machine gets up.
|
||||
|
||||
After the master machine gets up, we can query the public DNS of the master machine using the following command.
|
||||
```bash
|
||||
./yarn-ec2 -k mykey -i mypem.pem get-master xgboost
|
||||
```
|
||||
It will show the public DNS of the master machine like ```ec2-xx-xx-xx.us-west-2.compute.amazonaws.com```
|
||||
Now we can open the browser, and type (replace the DNS with the master DNS)
|
||||
```
|
||||
ec2-xx-xx-xx.us-west-2.compute.amazonaws.com:8088
|
||||
```
|
||||
This will show the job tracker of the YARN cluster. Note that we may have to wait a few minutes before the master finishes bootstrapping and starts the
|
||||
job tracker.
|
||||
|
||||
After the master machine gets up, we can freely add more slave machines to the cluster.
|
||||
The following command add m3.xlarge instances to the cluster.
|
||||
```bash
|
||||
./yarn-ec2 -k mykey -i mypem.pem -t m3.xlarge -s 2 addslave xgboost
|
||||
```
|
||||
We can also choose to add two spot instances
|
||||
```bash
|
||||
./yarn-ec2 -k mykey -i mypem.pem -t m3.xlarge -s 2 addspot xgboost
|
||||
```
|
||||
The slave machines will start up, bootstrap and report to the master.
|
||||
You can check if the slave machines are connected by clicking on the Nodes link on the job tracker.
|
||||
Or simply type the following URL (replace DNS ith the master DNS)
|
||||
```
|
||||
ec2-xx-xx-xx.us-west-2.compute.amazonaws.com:8088/cluster/nodes
|
||||
```
|
||||
|
||||
One thing we should note is that not all the links in the job tracker work.
|
||||
This is due to that many of them use the private IP of AWS, which can only be accessed by EC2.
|
||||
We can use ssh proxy to access these packages.
|
||||
Now that we have set up a cluster with one master and two slaves, we are ready to run the experiment.
|
||||
|
||||
|
||||
Build XGBoost with S3
|
||||
---------------------
|
||||
We can log into the master machine by the following command.
|
||||
```bash
|
||||
./yarn-ec2 -k mykey -i mypem.pem login xgboost
|
||||
```
|
||||
|
||||
We will be using S3 to host the data and the result model, so the data won't get lost after the cluster shutdown.
|
||||
To do so, we will need to build xgboost with S3 support. The only thing we need to do is to set ```USE_S3```
|
||||
variable to be true. This can be achieved by the following command.
|
||||
|
||||
```bash
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost
|
||||
cp make/config.mk config.mk
|
||||
echo "USE_S3=1" >> config.mk
|
||||
make -j4
|
||||
```
|
||||
Now we have built the XGBoost with S3 support. You can also enable HDFS support if you plan to store data on HDFS by turning on ```USE_HDFS``` option.
|
||||
|
||||
XGBoost also relies on the environment variable to access S3, so you will need to add the following two lines to `~/.bashrc` (replacing the strings with the correct ones)
|
||||
on the master machine as well.
|
||||
|
||||
```bash
|
||||
export AWS_ACCESS_KEY_ID=AKIAIOSFODNN7EXAMPLE
|
||||
export AWS_SECRET_ACCESS_KEY=wJalrXUtnFEMI/K7MDENG/bPxRfiCYEXAMPLEKEY
|
||||
export BUCKET=mybucket
|
||||
```
|
||||
|
||||
Host the Data on S3
|
||||
-------------------
|
||||
In this example, we will copy the example dataset in xgboost to the S3 bucket as input.
|
||||
In normal usecases, the dataset is usually created from existing distributed processing pipeline.
|
||||
We can use [s3cmd](http://s3tools.org/s3cmd) to copy the data into mybucket (replace ${BUCKET} with the real bucket name).
|
||||
|
||||
```bash
|
||||
cd xgboost
|
||||
s3cmd put demo/data/agaricus.txt.train s3://${BUCKET}/xgb-demo/train/
|
||||
s3cmd put demo/data/agaricus.txt.test s3://${BUCKET}/xgb-demo/test/
|
||||
```
|
||||
|
||||
Submit the Jobs
|
||||
---------------
|
||||
Now everything is ready, we can submit the xgboost distributed job to the YARN cluster.
|
||||
We will use the [dmlc-submit](https://github.com/dmlc/dmlc-core/tree/master/tracker) script to submit the job.
|
||||
|
||||
Now we can run the following script in the distributed training folder (replace ${BUCKET} with the real bucket name)
|
||||
```bash
|
||||
cd xgboost/demo/distributed-training
|
||||
# Use dmlc-submit to submit the job.
|
||||
../../dmlc-core/tracker/dmlc-submit --cluster=yarn --num-workers=2 --worker-cores=2\
|
||||
../../xgboost mushroom.aws.conf nthread=2\
|
||||
data=s3://${BUCKET}/xgb-demo/train\
|
||||
eval[test]=s3://${BUCKET}/xgb-demo/test\
|
||||
model_dir=s3://${BUCKET}/xgb-demo/model
|
||||
```
|
||||
All the configurations such as ```data``` and ```model_dir``` can also be directly written into the configuration file.
|
||||
Note that we only specified the folder path to the file, instead of the file name.
|
||||
XGBoost will read in all the files in that folder as the training and evaluation data.
|
||||
|
||||
In this command, we are using two workers, and each worker uses two running threads.
|
||||
XGBoost can benefit from using multiple cores in each worker.
|
||||
A common choice of working cores can range from 4 to 8.
|
||||
The trained model will be saved into the specified model folder. You can browse the model folder.
|
||||
```
|
||||
s3cmd ls s3://${BUCKET}/xgb-demo/model/
|
||||
```
|
||||
|
||||
The following is an example output from distributed training.
|
||||
```
|
||||
16/02/26 05:41:59 INFO dmlc.Client: jobname=DMLC[nworker=2]:xgboost,username=ubuntu
|
||||
16/02/26 05:41:59 INFO dmlc.Client: Submitting application application_1456461717456_0015
|
||||
16/02/26 05:41:59 INFO impl.YarnClientImpl: Submitted application application_1456461717456_0015
|
||||
2016-02-26 05:42:05,230 INFO @tracker All of 2 nodes getting started
|
||||
2016-02-26 05:42:14,027 INFO [05:42:14] [0] test-error:0.016139 train-error:0.014433
|
||||
2016-02-26 05:42:14,186 INFO [05:42:14] [1] test-error:0.000000 train-error:0.001228
|
||||
2016-02-26 05:42:14,947 INFO @tracker All nodes finishes job
|
||||
2016-02-26 05:42:14,948 INFO @tracker 9.71754479408 secs between node start and job finish
|
||||
Application application_1456461717456_0015 finished with state FINISHED at 1456465335961
|
||||
```
|
||||
|
||||
Analyze the Model
|
||||
-----------------
|
||||
After the model is trained, we can analyse the learnt model and use it for future prediction tasks.
|
||||
XGBoost is a portable framework, meaning the models in all platforms are ***exchangeable***.
|
||||
This means we can load the trained model in python/R/Julia and take benefit of data science pipelines
|
||||
in these languages to do model analysis and prediction.
|
||||
|
||||
For example, you can use [this IPython notebook](https://github.com/dmlc/xgboost/tree/master/demo/distributed-training/plot_model.ipynb)
|
||||
to plot feature importance and visualize the learnt model.
|
||||
|
||||
Troubleshooting
|
||||
----------------
|
||||
|
||||
If you encounter a problem, the best way might be to use the following command
|
||||
to get logs of stdout and stderr of the containers and check what causes the problem.
|
||||
```
|
||||
yarn logs -applicationId yourAppId
|
||||
```
|
||||
|
||||
Future Directions
|
||||
-----------------
|
||||
You have learned to use distributed XGBoost on YARN in this tutorial.
|
||||
XGBoost is a portable and scalable framework for gradient boosting.
|
||||
You can check out more examples and resources in the [resources page](https://github.com/dmlc/xgboost/blob/master/demo/README.md).
|
||||
|
||||
The project goal is to make the best scalable machine learning solution available to all platforms.
|
||||
The API is designed to be able to portable, and the same code can also run on other platforms such as MPI and SGE.
|
||||
XGBoost is actively evolving and we are working on even more exciting features
|
||||
such as distributed xgboost python/R package. Checkout [RoadMap](https://github.com/dmlc/xgboost/issues/873) for
|
||||
more details and you are more than welcomed to contribute to the project.
|
||||
216
doc/tutorials/aws_yarn.rst
Normal file
216
doc/tutorials/aws_yarn.rst
Normal file
@ -0,0 +1,216 @@
|
||||
###############################
|
||||
Distributed XGBoost YARN on AWS
|
||||
###############################
|
||||
This is a step-by-step tutorial on how to setup and run distributed `XGBoost <https://github.com/dmlc/xgboost>`_
|
||||
on an AWS EC2 cluster. Distributed XGBoost runs on various platforms such as MPI, SGE and Hadoop YARN.
|
||||
In this tutorial, we use YARN as an example since this is a widely used solution for distributed computing.
|
||||
|
||||
.. note:: XGBoost on Spark
|
||||
|
||||
If you are preprocessing training data with Spark, you may want to look at `XGBoost4J-Spark <https://xgboost.ai/2016/10/26/a-full-integration-of-xgboost-and-spark.html>`_, which supports distributed training on Resilient Distributed Dataset (RDD).
|
||||
|
||||
************
|
||||
Prerequisite
|
||||
************
|
||||
We need to get a `AWS key-pair <http://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ec2-key-pairs.html>`_
|
||||
to access the AWS services. Let us assume that we are using a key ``mykey`` and the corresponding permission file ``mypem.pem``.
|
||||
|
||||
We also need `AWS credentials <https://docs.aws.amazon.com/cli/latest/userguide/cli-chap-getting-started.html>`_,
|
||||
which includes an ``ACCESS_KEY_ID`` and a ``SECRET_ACCESS_KEY``.
|
||||
|
||||
Finally, we will need a S3 bucket to host the data and the model, ``s3://mybucket/``
|
||||
|
||||
***************************
|
||||
Setup a Hadoop YARN Cluster
|
||||
***************************
|
||||
This sections shows how to start a Hadoop YARN cluster from scratch.
|
||||
You can skip this step if you have already have one.
|
||||
We will be using `yarn-ec2 <https://github.com/tqchen/yarn-ec2>`_ to start the cluster.
|
||||
|
||||
We can first clone the yarn-ec2 script by the following command.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone https://github.com/tqchen/yarn-ec2
|
||||
|
||||
To use the script, we must set the environment variables ``AWS_ACCESS_KEY_ID`` and
|
||||
``AWS_SECRET_ACCESS_KEY`` properly. This can be done by adding the following two lines in
|
||||
``~/.bashrc`` (replacing the strings with the correct ones)
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
export AWS_ACCESS_KEY_ID=[your access ID]
|
||||
export AWS_SECRET_ACCESS_KEY=[your secret access key]
|
||||
|
||||
Now we can launch a master machine of the cluster from EC2:
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
./yarn-ec2 -k mykey -i mypem.pem launch xgboost
|
||||
|
||||
Wait a few mininutes till the master machine gets up.
|
||||
|
||||
After the master machine gets up, we can query the public DNS of the master machine using the following command.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
./yarn-ec2 -k mykey -i mypem.pem get-master xgboost
|
||||
|
||||
It will show the public DNS of the master machine like ``ec2-xx-xx-xx.us-west-2.compute.amazonaws.com``
|
||||
Now we can open the browser, and type (replace the DNS with the master DNS)
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
ec2-xx-xx-xx.us-west-2.compute.amazonaws.com:8088
|
||||
|
||||
This will show the job tracker of the YARN cluster. Note that we may have to wait a few minutes before the master finishes bootstrapping and starts the
|
||||
job tracker.
|
||||
|
||||
After the master machine gets up, we can freely add more slave machines to the cluster.
|
||||
The following command add m3.xlarge instances to the cluster.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
./yarn-ec2 -k mykey -i mypem.pem -t m3.xlarge -s 2 addslave xgboost
|
||||
|
||||
We can also choose to add two spot instances
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
./yarn-ec2 -k mykey -i mypem.pem -t m3.xlarge -s 2 addspot xgboost
|
||||
|
||||
The slave machines will start up, bootstrap and report to the master.
|
||||
You can check if the slave machines are connected by clicking on the Nodes link on the job tracker.
|
||||
Or simply type the following URL (replace DNS ith the master DNS)
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
ec2-xx-xx-xx.us-west-2.compute.amazonaws.com:8088/cluster/nodes
|
||||
|
||||
One thing we should note is that not all the links in the job tracker work.
|
||||
This is due to that many of them use the private IP of AWS, which can only be accessed by EC2.
|
||||
We can use ssh proxy to access these packages.
|
||||
Now that we have set up a cluster with one master and two slaves, we are ready to run the experiment.
|
||||
|
||||
*********************
|
||||
Build XGBoost with S3
|
||||
*********************
|
||||
We can log into the master machine by the following command.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
./yarn-ec2 -k mykey -i mypem.pem login xgboost
|
||||
|
||||
We will be using S3 to host the data and the result model, so the data won't get lost after the cluster shutdown.
|
||||
To do so, we will need to build XGBoost with S3 support. The only thing we need to do is to set ``USE_S3``
|
||||
variable to be true. This can be achieved by the following command.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
git clone --recursive https://github.com/dmlc/xgboost
|
||||
cd xgboost
|
||||
cp make/config.mk config.mk
|
||||
echo "USE_S3=1" >> config.mk
|
||||
make -j4
|
||||
|
||||
Now we have built the XGBoost with S3 support. You can also enable HDFS support if you plan to store data on HDFS by turning on ``USE_HDFS`` option.
|
||||
XGBoost also relies on the environment variable to access S3, so you will need to add the following two lines to ``~/.bashrc`` (replacing the strings with the correct ones)
|
||||
on the master machine as well.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
export AWS_ACCESS_KEY_ID=AKIAIOSFODNN7EXAMPLE
|
||||
export AWS_SECRET_ACCESS_KEY=wJalrXUtnFEMI/K7MDENG/bPxRfiCYEXAMPLEKEY
|
||||
export BUCKET=mybucket
|
||||
|
||||
*******************
|
||||
Host the Data on S3
|
||||
*******************
|
||||
In this example, we will copy the example dataset in XGBoost to the S3 bucket as input.
|
||||
In normal usecases, the dataset is usually created from existing distributed processing pipeline.
|
||||
We can use `s3cmd <http://s3tools.org/s3cmd>`_ to copy the data into mybucket (replace ``${BUCKET}`` with the real bucket name).
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cd xgboost
|
||||
s3cmd put demo/data/agaricus.txt.train s3://${BUCKET}/xgb-demo/train/
|
||||
s3cmd put demo/data/agaricus.txt.test s3://${BUCKET}/xgb-demo/test/
|
||||
|
||||
***************
|
||||
Submit the Jobs
|
||||
***************
|
||||
Now everything is ready, we can submit the XGBoost distributed job to the YARN cluster.
|
||||
We will use the `dmlc-submit <https://github.com/dmlc/dmlc-core/tree/master/tracker>`_ script to submit the job.
|
||||
|
||||
Now we can run the following script in the distributed training folder (replace ``${BUCKET}`` with the real bucket name)
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
cd xgboost/demo/distributed-training
|
||||
# Use dmlc-submit to submit the job.
|
||||
../../dmlc-core/tracker/dmlc-submit --cluster=yarn --num-workers=2 --worker-cores=2\
|
||||
../../xgboost mushroom.aws.conf nthread=2\
|
||||
data=s3://${BUCKET}/xgb-demo/train\
|
||||
eval[test]=s3://${BUCKET}/xgb-demo/test\
|
||||
model_dir=s3://${BUCKET}/xgb-demo/model
|
||||
|
||||
All the configurations such as ``data`` and ``model_dir`` can also be directly written into the configuration file.
|
||||
Note that we only specified the folder path to the file, instead of the file name.
|
||||
XGBoost will read in all the files in that folder as the training and evaluation data.
|
||||
|
||||
In this command, we are using two workers, and each worker uses two running threads.
|
||||
XGBoost can benefit from using multiple cores in each worker.
|
||||
A common choice of working cores can range from 4 to 8.
|
||||
The trained model will be saved into the specified model folder. You can browse the model folder.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
s3cmd ls s3://${BUCKET}/xgb-demo/model/
|
||||
|
||||
The following is an example output from distributed training.
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
16/02/26 05:41:59 INFO dmlc.Client: jobname=DMLC[nworker=2]:xgboost,username=ubuntu
|
||||
16/02/26 05:41:59 INFO dmlc.Client: Submitting application application_1456461717456_0015
|
||||
16/02/26 05:41:59 INFO impl.YarnClientImpl: Submitted application application_1456461717456_0015
|
||||
2016-02-26 05:42:05,230 INFO @tracker All of 2 nodes getting started
|
||||
2016-02-26 05:42:14,027 INFO [05:42:14] [0] test-error:0.016139 train-error:0.014433
|
||||
2016-02-26 05:42:14,186 INFO [05:42:14] [1] test-error:0.000000 train-error:0.001228
|
||||
2016-02-26 05:42:14,947 INFO @tracker All nodes finishes job
|
||||
2016-02-26 05:42:14,948 INFO @tracker 9.71754479408 secs between node start and job finish
|
||||
Application application_1456461717456_0015 finished with state FINISHED at 1456465335961
|
||||
|
||||
*****************
|
||||
Analyze the Model
|
||||
*****************
|
||||
After the model is trained, we can analyse the learnt model and use it for future prediction tasks.
|
||||
XGBoost is a portable framework, meaning the models in all platforms are *exchangeable*.
|
||||
This means we can load the trained model in python/R/Julia and take benefit of data science pipelines
|
||||
in these languages to do model analysis and prediction.
|
||||
|
||||
For example, you can use `this IPython notebook <https://github.com/dmlc/xgboost/tree/master/demo/distributed-training/plot_model.ipynb>`_
|
||||
to plot feature importance and visualize the learnt model.
|
||||
|
||||
***************
|
||||
Troubleshooting
|
||||
***************
|
||||
|
||||
If you encounter a problem, the best way might be to use the following command
|
||||
to get logs of stdout and stderr of the containers and check what causes the problem.
|
||||
|
||||
.. code-block:: bash
|
||||
|
||||
yarn logs -applicationId yourAppId
|
||||
|
||||
*****************
|
||||
Future Directions
|
||||
*****************
|
||||
You have learned to use distributed XGBoost on YARN in this tutorial.
|
||||
XGBoost is a portable and scalable framework for gradient boosting.
|
||||
You can check out more examples and resources in the `resources page <https://github.com/dmlc/xgboost/blob/master/demo/README.md>`_.
|
||||
|
||||
The project goal is to make the best scalable machine learning solution available to all platforms.
|
||||
The API is designed to be able to portable, and the same code can also run on other platforms such as MPI and SGE.
|
||||
XGBoost is actively evolving and we are working on even more exciting features
|
||||
such as distributed XGBoost python/R package.
|
||||
@ -1,101 +0,0 @@
|
||||
DART booster
|
||||
============
|
||||
[XGBoost](https://github.com/dmlc/xgboost) mostly combines a huge number of regression trees with a small learning rate.
|
||||
In this situation, trees added early are significant and trees added late are unimportant.
|
||||
|
||||
Vinayak and Gilad-Bachrach proposed a new method to add dropout techniques from the deep neural net community to boosted trees, and reported better results in some situations.
|
||||
|
||||
This is a instruction of new tree booster `dart`.
|
||||
|
||||
Original paper
|
||||
--------------
|
||||
Rashmi Korlakai Vinayak, Ran Gilad-Bachrach. "DART: Dropouts meet Multiple Additive Regression Trees." [JMLR](http://www.jmlr.org/proceedings/papers/v38/korlakaivinayak15.pdf)
|
||||
|
||||
Features
|
||||
--------
|
||||
- Drop trees in order to solve the over-fitting.
|
||||
- Trivial trees (to correct trivial errors) may be prevented.
|
||||
|
||||
Because of the randomness introduced in the training, expect the following few differences:
|
||||
- Training can be slower than `gbtree` because the random dropout prevents usage of the prediction buffer.
|
||||
- The early stop might not be stable, due to the randomness.
|
||||
|
||||
How it works
|
||||
------------
|
||||
- In ``$ m $``th training round, suppose ``$ k $`` trees are selected to be dropped.
|
||||
- Let ``$ D = \sum_{i \in \mathbf{K}} F_i $`` be the leaf scores of dropped trees and ``$ F_m = \eta \tilde{F}_m $`` be the leaf scores of a new tree.
|
||||
- The objective function is as follows:
|
||||
```math
|
||||
\mathrm{Obj}
|
||||
= \sum_{j=1}^n L \left( y_j, \hat{y}_j^{m-1} - D_j + \tilde{F}_m \right)
|
||||
+ \Omega \left( \tilde{F}_m \right).
|
||||
```
|
||||
- ``$ D $`` and ``$ F_m $`` are overshooting, so using scale factor
|
||||
```math
|
||||
\hat{y}_j^m = \sum_{i \not\in \mathbf{K}} F_i + a \left( \sum_{i \in \mathbf{K}} F_i + b F_m \right) .
|
||||
```
|
||||
|
||||
Parameters
|
||||
----------
|
||||
### booster
|
||||
* `dart`
|
||||
|
||||
This booster inherits `gbtree`, so `dart` has also `eta`, `gamma`, `max_depth` and so on.
|
||||
|
||||
Additional parameters are noted below.
|
||||
|
||||
### sample_type
|
||||
type of sampling algorithm.
|
||||
* `uniform`: (default) dropped trees are selected uniformly.
|
||||
* `weighted`: dropped trees are selected in proportion to weight.
|
||||
|
||||
### normalize_type
|
||||
type of normalization algorithm.
|
||||
* `tree`: (default) New trees have the same weight of each of dropped trees.
|
||||
```math
|
||||
a \left( \sum_{i \in \mathbf{K}} F_i + \frac{1}{k} F_m \right)
|
||||
&= a \left( \sum_{i \in \mathbf{K}} F_i + \frac{\eta}{k} \tilde{F}_m \right) \\
|
||||
&\sim a \left( 1 + \frac{\eta}{k} \right) D \\
|
||||
&= a \frac{k + \eta}{k} D = D , \\
|
||||
&\quad a = \frac{k}{k + \eta} .
|
||||
```
|
||||
|
||||
* `forest`: New trees have the same weight of sum of dropped trees (forest).
|
||||
```math
|
||||
a \left( \sum_{i \in \mathbf{K}} F_i + F_m \right)
|
||||
&= a \left( \sum_{i \in \mathbf{K}} F_i + \eta \tilde{F}_m \right) \\
|
||||
&\sim a \left( 1 + \eta \right) D \\
|
||||
&= a (1 + \eta) D = D , \\
|
||||
&\quad a = \frac{1}{1 + \eta} .
|
||||
```
|
||||
|
||||
### rate_drop
|
||||
dropout rate.
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
### skip_drop
|
||||
probability of skipping dropout.
|
||||
- If a dropout is skipped, new trees are added in the same manner as gbtree.
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
Sample Script
|
||||
-------------
|
||||
```python
|
||||
import xgboost as xgb
|
||||
# read in data
|
||||
dtrain = xgb.DMatrix('demo/data/agaricus.txt.train')
|
||||
dtest = xgb.DMatrix('demo/data/agaricus.txt.test')
|
||||
# specify parameters via map
|
||||
param = {'booster': 'dart',
|
||||
'max_depth': 5, 'learning_rate': 0.1,
|
||||
'objective': 'binary:logistic', 'silent': True,
|
||||
'sample_type': 'uniform',
|
||||
'normalize_type': 'tree',
|
||||
'rate_drop': 0.1,
|
||||
'skip_drop': 0.5}
|
||||
num_round = 50
|
||||
bst = xgb.train(param, dtrain, num_round)
|
||||
# make prediction
|
||||
# ntree_limit must not be 0
|
||||
preds = bst.predict(dtest, ntree_limit=num_round)
|
||||
```
|
||||
113
doc/tutorials/dart.rst
Normal file
113
doc/tutorials/dart.rst
Normal file
@ -0,0 +1,113 @@
|
||||
############
|
||||
DART booster
|
||||
############
|
||||
XGBoost mostly combines a huge number of regression trees with a small learning rate.
|
||||
In this situation, trees added early are significant and trees added late are unimportant.
|
||||
|
||||
Vinayak and Gilad-Bachrach proposed a new method to add dropout techniques from the deep neural net community to boosted trees, and reported better results in some situations.
|
||||
|
||||
This is a instruction of new tree booster ``dart``.
|
||||
|
||||
**************
|
||||
Original paper
|
||||
**************
|
||||
Rashmi Korlakai Vinayak, Ran Gilad-Bachrach. "DART: Dropouts meet Multiple Additive Regression Trees." `JMLR <http://www.jmlr.org/proceedings/papers/v38/korlakaivinayak15.pdf>`_.
|
||||
|
||||
********
|
||||
Features
|
||||
********
|
||||
- Drop trees in order to solve the over-fitting.
|
||||
|
||||
- Trivial trees (to correct trivial errors) may be prevented.
|
||||
|
||||
Because of the randomness introduced in the training, expect the following few differences:
|
||||
|
||||
- Training can be slower than ``gbtree`` because the random dropout prevents usage of the prediction buffer.
|
||||
- The early stop might not be stable, due to the randomness.
|
||||
|
||||
************
|
||||
How it works
|
||||
************
|
||||
- In :math:`m`-th training round, suppose :math:`k` trees are selected to be dropped.
|
||||
- Let :math:`D = \sum_{i \in \mathbf{K}} F_i` be the leaf scores of dropped trees and :math:`F_m = \eta \tilde{F}_m` be the leaf scores of a new tree.
|
||||
- The objective function is as follows:
|
||||
|
||||
.. math::
|
||||
|
||||
\mathrm{Obj}
|
||||
= \sum_{j=1}^n L \left( y_j, \hat{y}_j^{m-1} - D_j + \tilde{F}_m \right)
|
||||
+ \Omega \left( \tilde{F}_m \right).
|
||||
|
||||
- :math:`D` and :math:`F_m` are overshooting, so using scale factor
|
||||
|
||||
.. math::
|
||||
|
||||
\hat{y}_j^m = \sum_{i \not\in \mathbf{K}} F_i + a \left( \sum_{i \in \mathbf{K}} F_i + b F_m \right) .
|
||||
|
||||
**********
|
||||
Parameters
|
||||
**********
|
||||
|
||||
The booster ``dart`` inherits ``gbtree`` booster, so it supports all parameters that ``gbtree`` does, such as ``eta``, ``gamma``, ``max_depth`` etc.
|
||||
|
||||
Additional parameters are noted below:
|
||||
|
||||
* ``sample_type``: type of sampling algorithm.
|
||||
|
||||
- ``uniform``: (default) dropped trees are selected uniformly.
|
||||
- ``weighted``: dropped trees are selected in proportion to weight.
|
||||
|
||||
* ``normalize_type``: type of normalization algorithm.
|
||||
|
||||
- ``tree``: (default) New trees have the same weight of each of dropped trees.
|
||||
|
||||
.. math::
|
||||
|
||||
a \left( \sum_{i \in \mathbf{K}} F_i + \frac{1}{k} F_m \right)
|
||||
&= a \left( \sum_{i \in \mathbf{K}} F_i + \frac{\eta}{k} \tilde{F}_m \right) \\
|
||||
&\sim a \left( 1 + \frac{\eta}{k} \right) D \\
|
||||
&= a \frac{k + \eta}{k} D = D , \\
|
||||
&\quad a = \frac{k}{k + \eta}
|
||||
|
||||
- ``forest``: New trees have the same weight of sum of dropped trees (forest).
|
||||
|
||||
.. math::
|
||||
|
||||
a \left( \sum_{i \in \mathbf{K}} F_i + F_m \right)
|
||||
&= a \left( \sum_{i \in \mathbf{K}} F_i + \eta \tilde{F}_m \right) \\
|
||||
&\sim a \left( 1 + \eta \right) D \\
|
||||
&= a (1 + \eta) D = D , \\
|
||||
&\quad a = \frac{1}{1 + \eta} .
|
||||
|
||||
* ``rate_drop``: dropout rate.
|
||||
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
* ``skip_drop``: probability of skipping dropout.
|
||||
|
||||
- If a dropout is skipped, new trees are added in the same manner as gbtree.
|
||||
- range: [0.0, 1.0]
|
||||
|
||||
*************
|
||||
Sample Script
|
||||
*************
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
import xgboost as xgb
|
||||
# read in data
|
||||
dtrain = xgb.DMatrix('demo/data/agaricus.txt.train')
|
||||
dtest = xgb.DMatrix('demo/data/agaricus.txt.test')
|
||||
# specify parameters via map
|
||||
param = {'booster': 'dart',
|
||||
'max_depth': 5, 'learning_rate': 0.1,
|
||||
'objective': 'binary:logistic', 'silent': True,
|
||||
'sample_type': 'uniform',
|
||||
'normalize_type': 'tree',
|
||||
'rate_drop': 0.1,
|
||||
'skip_drop': 0.5}
|
||||
num_round = 50
|
||||
bst = xgb.train(param, dtrain, num_round)
|
||||
# make prediction
|
||||
# ntree_limit must not be 0
|
||||
preds = bst.predict(dtest, ntree_limit=num_round)
|
||||
51
doc/tutorials/external_memory.rst
Normal file
51
doc/tutorials/external_memory.rst
Normal file
@ -0,0 +1,51 @@
|
||||
############################################
|
||||
Using XGBoost External Memory Version (beta)
|
||||
############################################
|
||||
There is no big difference between using external memory version and in-memory version.
|
||||
The only difference is the filename format.
|
||||
|
||||
The external memory version takes in the following filename format:
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
filename#cacheprefix
|
||||
|
||||
The ``filename`` is the normal path to libsvm file you want to load in, and ``cacheprefix`` is a
|
||||
path to a cache file that XGBoost will use for external memory cache.
|
||||
|
||||
The following code was extracted from `demo/guide-python/external_memory.py <https://github.com/dmlc/xgboost/blob/master/demo/guide-python/external_memory.py>`_:
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
dtrain = xgb.DMatrix('../data/agaricus.txt.train#dtrain.cache')
|
||||
|
||||
You can find that there is additional ``#dtrain.cache`` following the libsvm file, this is the name of cache file.
|
||||
For CLI version, simply add the cache suffix, e.g. ``"../data/agaricus.txt.train#dtrain.cache"``.
|
||||
|
||||
****************
|
||||
Performance Note
|
||||
****************
|
||||
* the parameter ``nthread`` should be set to number of **physical** cores
|
||||
|
||||
- Most modern CPUs use hyperthreading, which means a 4 core CPU may carry 8 threads
|
||||
- Set ``nthread`` to be 4 for maximum performance in such case
|
||||
|
||||
*******************
|
||||
Distributed Version
|
||||
*******************
|
||||
The external memory mode naturally works on distributed version, you can simply set path like
|
||||
|
||||
.. code-block:: none
|
||||
|
||||
data = "hdfs://path-to-data/#dtrain.cache"
|
||||
|
||||
XGBoost will cache the data to the local position. When you run on YARN, the current folder is temporal
|
||||
so that you can directly use ``dtrain.cache`` to cache to current folder.
|
||||
|
||||
**********
|
||||
Usage Note
|
||||
**********
|
||||
* This is a experimental version
|
||||
* Currently only importing from libsvm format is supported
|
||||
|
||||
- Contribution of ingestion from other common external memory data source is welcomed
|
||||
@ -1,10 +0,0 @@
|
||||
# XGBoost Tutorials
|
||||
|
||||
This section contains official tutorials inside XGBoost package.
|
||||
See [Awesome XGBoost](https://github.com/dmlc/xgboost/tree/master/demo) for links to more resources.
|
||||
|
||||
## Contents
|
||||
- [Introduction to Boosted Trees](../model.md)
|
||||
- [Distributed XGBoost YARN on AWS](aws_yarn.md)
|
||||
- [DART booster](dart.md)
|
||||
- [Monotonic Constraints](monotonic.md)
|
||||
19
doc/tutorials/index.rst
Normal file
19
doc/tutorials/index.rst
Normal file
@ -0,0 +1,19 @@
|
||||
#################
|
||||
XGBoost Tutorials
|
||||
#################
|
||||
|
||||
This section contains official tutorials inside XGBoost package.
|
||||
See `Awesome XGBoost <https://github.com/dmlc/xgboost/tree/master/demo>`_ for more resources.
|
||||
|
||||
.. toctree::
|
||||
:maxdepth: 1
|
||||
:caption: Contents:
|
||||
|
||||
model
|
||||
aws_yarn
|
||||
dart
|
||||
monotonic
|
||||
input_format
|
||||
param_tuning
|
||||
external_memory
|
||||
|
||||
112
doc/tutorials/input_format.rst
Normal file
112
doc/tutorials/input_format.rst
Normal file
@ -0,0 +1,112 @@
|
||||
############################
|
||||
Text Input Format of DMatrix
|
||||
############################
|
||||
|
||||
******************
|
||||
Basic Input Format
|
||||
******************
|
||||
XGBoost currently supports two text formats for ingesting data: LibSVM and CSV. The rest of this document will describe the LibSVM format. (See `this Wikipedia article <https://en.wikipedia.org/wiki/Comma-separated_values>`_ for a description of the CSV format.)
|
||||
|
||||
For training or predicting, XGBoost takes an instance file with the format as below:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt``
|
||||
|
||||
1 101:1.2 102:0.03
|
||||
0 1:2.1 10001:300 10002:400
|
||||
0 0:1.3 1:0.3
|
||||
1 0:0.01 1:0.3
|
||||
0 0:0.2 1:0.3
|
||||
|
||||
Each line represent a single instance, and in the first line '1' is the instance label, '101' and '102' are feature indices, '1.2' and '0.03' are feature values. In the binary classification case, '1' is used to indicate positive samples, and '0' is used to indicate negative samples. We also support probability values in [0,1] as label, to indicate the probability of the instance being positive.
|
||||
|
||||
******************************************
|
||||
Auxiliary Files for Additional Information
|
||||
******************************************
|
||||
**Note: all information below is applicable only to single-node version of the package.** If you'd like to perform distributed training with multiple nodes, skip to the section `Embedding additional information inside LibSVM file`_.
|
||||
|
||||
Group Input Format
|
||||
==================
|
||||
For `ranking task <https://github.com/dmlc/xgboost/tree/master/demo/rank>`_, XGBoost supports the group input format. In ranking task, instances are categorized into *query groups* in real world scenarios. For example, in the learning to rank web pages scenario, the web page instances are grouped by their queries. XGBoost requires an file that indicates the group information. For example, if the instance file is the ``train.txt`` shown above, the group file should be named ``train.txt.group`` and be of the following format:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt.group``
|
||||
|
||||
2
|
||||
3
|
||||
|
||||
This means that, the data set contains 5 instances, and the first two instances are in a group and the other three are in another group. The numbers in the group file are actually indicating the number of instances in each group in the instance file in order.
|
||||
At the time of configuration, you do not have to indicate the path of the group file. If the instance file name is ``xxx``, XGBoost will check whether there is a file named ``xxx.group`` in the same directory.
|
||||
|
||||
Instance Weight File
|
||||
====================
|
||||
Instances in the training data may be assigned weights to differentiate relative importance among them. For example, if we provide an instance weight file for the ``train.txt`` file in the example as below:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt.weight``
|
||||
|
||||
1
|
||||
0.5
|
||||
0.5
|
||||
1
|
||||
0.5
|
||||
|
||||
It means that XGBoost will emphasize more on the first and fourth instance (i.e. the positive instances) while training.
|
||||
The configuration is similar to configuring the group information. If the instance file name is ``xxx``, XGBoost will look for a file named ``xxx.weight`` in the same directory. If the file exists, the instance weights will be extracted and used at the time of training.
|
||||
|
||||
.. note:: Binary buffer format and instance weights
|
||||
|
||||
If you choose to save the training data as a binary buffer (using :py:meth:`save_binary() <xgboost.DMatrix.save_binary>`), keep in mind that the resulting binary buffer file will include the instance weights. To update the weights, use the :py:meth:`set_weight() <xgboost.DMatrix.set_weight>` function.
|
||||
|
||||
Initial Margin File
|
||||
===================
|
||||
XGBoost supports providing each instance an initial margin prediction. For example, if we have a initial prediction using logistic regression for ``train.txt`` file, we can create the following file:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt.base_margin``
|
||||
|
||||
-0.4
|
||||
1.0
|
||||
3.4
|
||||
|
||||
XGBoost will take these values as initial margin prediction and boost from that. An important note about base_margin is that it should be margin prediction before transformation, so if you are doing logistic loss, you will need to put in value before logistic transformation. If you are using XGBoost predictor, use ``pred_margin=1`` to output margin values.
|
||||
|
||||
***************************************************
|
||||
Embedding additional information inside LibSVM file
|
||||
***************************************************
|
||||
**This section is applicable to both single- and multiple-node settings.**
|
||||
|
||||
Query ID Columns
|
||||
================
|
||||
This is most useful for `ranking task <https://github.com/dmlc/xgboost/tree/master/demo/rank>`_, where the instances are grouped into query groups. You may embed query group ID for each instance in the LibSVM file by adding a token of form ``qid:xx`` in each row:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt``
|
||||
|
||||
1 qid:1 101:1.2 102:0.03
|
||||
0 qid:1 1:2.1 10001:300 10002:400
|
||||
0 qid:2 0:1.3 1:0.3
|
||||
1 qid:2 0:0.01 1:0.3
|
||||
0 qid:3 0:0.2 1:0.3
|
||||
1 qid:3 3:-0.1 10:-0.3
|
||||
0 qid:3 6:0.2 10:0.15
|
||||
|
||||
Keep in mind the following restrictions:
|
||||
|
||||
* You are not allowed to specify query ID's for some instances but not for others. Either every row is assigned query ID's or none at all.
|
||||
* The rows have to be sorted in ascending order by the query IDs. So, for instance, you may not have one row having large query ID than any of the following rows.
|
||||
|
||||
Instance weights
|
||||
================
|
||||
You may specify instance weights in the LibSVM file by appending each instance label with the corresponding weight in the form of ``[label]:[weight]``, as shown by the following example:
|
||||
|
||||
.. code-block:: none
|
||||
:caption: ``train.txt``
|
||||
|
||||
1:1.0 101:1.2 102:0.03
|
||||
0:0.5 1:2.1 10001:300 10002:400
|
||||
0:0.5 0:1.3 1:0.3
|
||||
1:1.0 0:0.01 1:0.3
|
||||
0:0.5 0:0.2 1:0.3
|
||||
|
||||
where the negative instances are assigned half weights compared to the positive instances.
|
||||
265
doc/tutorials/model.rst
Normal file
265
doc/tutorials/model.rst
Normal file
@ -0,0 +1,265 @@
|
||||
#############################
|
||||
Introduction to Boosted Trees
|
||||
#############################
|
||||
XGBoost stands for "Extreme Gradient Boosting", where the term "Gradient Boosting" originates from the paper *Greedy Function Approximation: A Gradient Boosting Machine*, by Friedman.
|
||||
This is a tutorial on gradient boosted trees, and most of the content is based on `these slides <http://homes.cs.washington.edu/~tqchen/pdf/BoostedTree.pdf>`_ by Tianqi Chen, the original author of XGBoost.
|
||||
|
||||
The **gradient boosted trees** has been around for a while, and there are a lot of materials on the topic.
|
||||
This tutorial will explain boosted trees in a self-contained and principled way using the elements of supervised learning.
|
||||
We think this explanation is cleaner, more formal, and motivates the model formulation used in XGBoost.
|
||||
|
||||
*******************************
|
||||
Elements of Supervised Learning
|
||||
*******************************
|
||||
XGBoost is used for supervised learning problems, where we use the training data (with multiple features) :math:`x_i` to predict a target variable :math:`y_i`.
|
||||
Before we learn about trees specifically, let us start by reviewing the basic elements in supervised learning.
|
||||
|
||||
Model and Parameters
|
||||
====================
|
||||
The **model** in supervised learning usually refers to the mathematical structure of by which the prediction :math:`y_i` is made from the input :math:`x_i`.
|
||||
A common example is a *linear model*, where the prediction is given as :math:`\hat{y}_i = \sum_j \theta_j x_{ij}`, a linear combination of weighted input features.
|
||||
The prediction value can have different interpretations, depending on the task, i.e., regression or classification.
|
||||
For example, it can be logistic transformed to get the probability of positive class in logistic regression, and it can also be used as a ranking score when we want to rank the outputs.
|
||||
|
||||
The **parameters** are the undetermined part that we need to learn from data. In linear regression problems, the parameters are the coefficients :math:`\theta`.
|
||||
Usually we will use :math:`\theta` to denote the parameters (there are many parameters in a model, our definition here is sloppy).
|
||||
|
||||
Objective Function: Training Loss + Regularization
|
||||
==================================================
|
||||
With judicious choices for :math:`y_i`, we may express a variety of tasks, such as regression, classification, and ranking.
|
||||
The task of **training** the model amounts to finding the best parameters :math:`\theta` that best fit the training data :math:`x_i` and labels :math:`y_i`. In order to train the model, we need to define the **objective function**
|
||||
to measure how well the model fit the training data.
|
||||
|
||||
A salient characteristic of objective functions is that they consist two parts: **training loss** and **regularization term**:
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}(\theta) = L(\theta) + \Omega(\theta)
|
||||
|
||||
where :math:`L` is the training loss function, and :math:`\Omega` is the regularization term. The training loss measures how *predictive* our model is with respect to the training data.
|
||||
A common choice of :math:`L` is the *mean squared error*, which is given by
|
||||
|
||||
.. math::
|
||||
|
||||
L(\theta) = \sum_i (y_i-\hat{y}_i)^2
|
||||
|
||||
Another commonly used loss function is logistic loss, to be used for logistic regression:
|
||||
|
||||
.. math::
|
||||
|
||||
L(\theta) = \sum_i[ y_i\ln (1+e^{-\hat{y}_i}) + (1-y_i)\ln (1+e^{\hat{y}_i})]
|
||||
|
||||
The **regularization term** is what people usually forget to add. The regularization term controls the complexity of the model, which helps us to avoid overfitting.
|
||||
This sounds a bit abstract, so let us consider the following problem in the following picture. You are asked to *fit* visually a step function given the input data points
|
||||
on the upper left corner of the image.
|
||||
Which solution among the three do you think is the best fit?
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/model/step_fit.png
|
||||
:alt: step functions to fit data points, illustrating bias-variance tradeoff
|
||||
|
||||
The correct answer is marked in red. Please consider if this visually seems a reasonable fit to you. The general principle is we want both a *simple* and *predictive* model.
|
||||
The tradeoff between the two is also referred as **bias-variance tradeoff** in machine learning.
|
||||
|
||||
Why introduce the general principle?
|
||||
====================================
|
||||
The elements introduced above form the basic elements of supervised learning, and they are natural building blocks of machine learning toolkits.
|
||||
For example, you should be able to describe the differences and commonalities between gradient boosted trees and random forests.
|
||||
Understanding the process in a formalized way also helps us to understand the objective that we are learning and the reason behind the heuristics such as
|
||||
pruning and smoothing.
|
||||
|
||||
***********************
|
||||
Decision Tree Ensembles
|
||||
***********************
|
||||
Now that we have introduced the elements of supervised learning, let us get started with real trees.
|
||||
To begin with, let us first learn about the model choice of XGBoost: **decision tree ensembles**.
|
||||
The tree ensemble model consists of a set of classification and regression trees (CART). Here's a simple example of a CART
|
||||
that classifies whether someone will like computer games.
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/model/cart.png
|
||||
:width: 100%
|
||||
:alt: a toy example for CART
|
||||
|
||||
We classify the members of a family into different leaves, and assign them the score on the corresponding leaf.
|
||||
A CART is a bit different from decision trees, in which the leaf only contains decision values. In CART, a real score
|
||||
is associated with each of the leaves, which gives us richer interpretations that go beyond classification.
|
||||
This also allows for a pricipled, unified approach to optimization, as we will see in a later part of this tutorial.
|
||||
|
||||
Usually, a single tree is not strong enough to be used in practice. What is actually used is the ensemble model,
|
||||
which sums the prediction of multiple trees together.
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/model/twocart.png
|
||||
:width: 100%
|
||||
:alt: a toy example for tree ensemble, consisting of two CARTs
|
||||
|
||||
Here is an example of a tree ensemble of two trees. The prediction scores of each individual tree are summed up to get the final score.
|
||||
If you look at the example, an important fact is that the two trees try to *complement* each other.
|
||||
Mathematically, we can write our model in the form
|
||||
|
||||
.. math::
|
||||
|
||||
\hat{y}_i = \sum_{k=1}^K f_k(x_i), f_k \in \mathcal{F}
|
||||
|
||||
where :math:`K` is the number of trees, :math:`f` is a function in the functional space :math:`\mathcal{F}`, and :math:`\mathcal{F}` is the set of all possible CARTs. The objective function to be optimized is given by
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}(\theta) = \sum_i^n l(y_i, \hat{y}_i) + \sum_{k=1}^K \Omega(f_k)
|
||||
|
||||
Now here comes a trick question: what is the *model* used in random forests? Tree ensembles! So random forests and boosted trees are really the same models; the
|
||||
difference arises from how we train them. This means that, if you write a predictive service for tree ensembles, you only need to write one and it should work
|
||||
for both random forests and gradient boosted trees. (See `Treelite <http://treelite.io>`_ for an actual example.) One example of why elements of supervised learning rock.
|
||||
|
||||
*************
|
||||
Tree Boosting
|
||||
*************
|
||||
Now that we introduced the model, let us turn to training: How should we learn the trees?
|
||||
The answer is, as is always for all supervised learning models: *define an objective function and optimize it*!
|
||||
|
||||
Let the following be the objective function (remember it always needs to contain training loss and regularization):
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj} = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t)}) + \sum_{i=1}^t\Omega(f_i)
|
||||
|
||||
Additive Training
|
||||
=================
|
||||
|
||||
The first question we want to ask: what are the **parameters** of trees?
|
||||
You can find that what we need to learn are those functions :math:`f_i`, each containing the structure
|
||||
of the tree and the leaf scores. Learning tree structure is much harder than traditional optimization problem where you can simply take the gradient.
|
||||
It is intractable to learn all the trees at once.
|
||||
Instead, we use an additive strategy: fix what we have learned, and add one new tree at a time.
|
||||
We write the prediction value at step :math:`t` as :math:`\hat{y}_i^{(t)}`. Then we have
|
||||
|
||||
.. math::
|
||||
|
||||
\hat{y}_i^{(0)} &= 0\\
|
||||
\hat{y}_i^{(1)} &= f_1(x_i) = \hat{y}_i^{(0)} + f_1(x_i)\\
|
||||
\hat{y}_i^{(2)} &= f_1(x_i) + f_2(x_i)= \hat{y}_i^{(1)} + f_2(x_i)\\
|
||||
&\dots\\
|
||||
\hat{y}_i^{(t)} &= \sum_{k=1}^t f_k(x_i)= \hat{y}_i^{(t-1)} + f_t(x_i)
|
||||
|
||||
It remains to ask: which tree do we want at each step? A natural thing is to add the one that optimizes our objective.
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}^{(t)} & = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t)}) + \sum_{i=1}^t\Omega(f_i) \\
|
||||
& = \sum_{i=1}^n l(y_i, \hat{y}_i^{(t-1)} + f_t(x_i)) + \Omega(f_t) + \mathrm{constant}
|
||||
|
||||
If we consider using mean squared error (MSE) as our loss function, the objective becomes
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}^{(t)} & = \sum_{i=1}^n (y_i - (\hat{y}_i^{(t-1)} + f_t(x_i)))^2 + \sum_{i=1}^t\Omega(f_i) \\
|
||||
& = \sum_{i=1}^n [2(\hat{y}_i^{(t-1)} - y_i)f_t(x_i) + f_t(x_i)^2] + \Omega(f_t) + \mathrm{constant}
|
||||
|
||||
The form of MSE is friendly, with a first order term (usually called the residual) and a quadratic term.
|
||||
For other losses of interest (for example, logistic loss), it is not so easy to get such a nice form.
|
||||
So in the general case, we take the *Taylor expansion of the loss function up to the second order*:
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}^{(t)} = \sum_{i=1}^n [l(y_i, \hat{y}_i^{(t-1)}) + g_i f_t(x_i) + \frac{1}{2} h_i f_t^2(x_i)] + \Omega(f_t) + \mathrm{constant}
|
||||
|
||||
where the :math:`g_i` and :math:`h_i` are defined as
|
||||
|
||||
.. math::
|
||||
|
||||
g_i &= \partial_{\hat{y}_i^{(t-1)}} l(y_i, \hat{y}_i^{(t-1)})\\
|
||||
h_i &= \partial_{\hat{y}_i^{(t-1)}}^2 l(y_i, \hat{y}_i^{(t-1)})
|
||||
|
||||
After we remove all the constants, the specific objective at step :math:`t` becomes
|
||||
|
||||
.. math::
|
||||
|
||||
\sum_{i=1}^n [g_i f_t(x_i) + \frac{1}{2} h_i f_t^2(x_i)] + \Omega(f_t)
|
||||
|
||||
This becomes our optimization goal for the new tree. One important advantage of this definition is that
|
||||
the value of the objective function only depends on :math:`g_i` and :math:`h_i`. This is how XGBoost supports custom loss functions.
|
||||
We can optimize every loss function, including logistic regression and pairwise ranking, using exactly
|
||||
the same solver that takes :math:`g_i` and :math:`h_i` as input!
|
||||
|
||||
Model Complexity
|
||||
================
|
||||
We have introduced the training step, but wait, there is one important thing, the **regularization term**!
|
||||
We need to define the complexity of the tree :math:`\Omega(f)`. In order to do so, let us first refine the definition of the tree :math:`f(x)` as
|
||||
|
||||
.. math::
|
||||
|
||||
f_t(x) = w_{q(x)}, w \in R^T, q:R^d\rightarrow \{1,2,\cdots,T\} .
|
||||
|
||||
Here :math:`w` is the vector of scores on leaves, :math:`q` is a function assigning each data point to the corresponding leaf, and :math:`T` is the number of leaves.
|
||||
In XGBoost, we define the complexity as
|
||||
|
||||
.. math::
|
||||
|
||||
\Omega(f) = \gamma T + \frac{1}{2}\lambda \sum_{j=1}^T w_j^2
|
||||
|
||||
Of course, there is more than one way to define the complexity, but this one works well in practice. The regularization is one part most tree packages treat
|
||||
less carefully, or simply ignore. This was because the traditional treatment of tree learning only emphasized improving impurity, while the complexity control was left to heuristics.
|
||||
By defining it formally, we can get a better idea of what we are learning and obtain models that perform well in the wild.
|
||||
|
||||
The Structure Score
|
||||
===================
|
||||
Here is the magical part of the derivation. After re-formulating the tree model, we can write the objective value with the :math:`t`-th tree as:
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}^{(t)} &\approx \sum_{i=1}^n [g_i w_{q(x_i)} + \frac{1}{2} h_i w_{q(x_i)}^2] + \gamma T + \frac{1}{2}\lambda \sum_{j=1}^T w_j^2\\
|
||||
&= \sum^T_{j=1} [(\sum_{i\in I_j} g_i) w_j + \frac{1}{2} (\sum_{i\in I_j} h_i + \lambda) w_j^2 ] + \gamma T
|
||||
|
||||
where :math:`I_j = \{i|q(x_i)=j\}` is the set of indices of data points assigned to the :math:`j`-th leaf.
|
||||
Notice that in the second line we have changed the index of the summation because all the data points on the same leaf get the same score.
|
||||
We could further compress the expression by defining :math:`G_j = \sum_{i\in I_j} g_i` and :math:`H_j = \sum_{i\in I_j} h_i`:
|
||||
|
||||
.. math::
|
||||
|
||||
\text{obj}^{(t)} = \sum^T_{j=1} [G_jw_j + \frac{1}{2} (H_j+\lambda) w_j^2] +\gamma T
|
||||
|
||||
In this equation, :math:`w_j` are independent with respect to each other, the form :math:`G_jw_j+\frac{1}{2}(H_j+\lambda)w_j^2` is quadratic and the best :math:`w_j` for a given structure :math:`q(x)` and the best objective reduction we can get is:
|
||||
|
||||
.. math::
|
||||
|
||||
w_j^\ast &= -\frac{G_j}{H_j+\lambda}\\
|
||||
\text{obj}^\ast &= -\frac{1}{2} \sum_{j=1}^T \frac{G_j^2}{H_j+\lambda} + \gamma T
|
||||
|
||||
The last equation measures *how good* a tree structure :math:`$q(x)` is.
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/model/struct_score.png
|
||||
:width: 100%
|
||||
:alt: illustration of structure score (fitness)
|
||||
|
||||
If all this sounds a bit complicated, let's take a look at the picture, and see how the scores can be calculated.
|
||||
Basically, for a given tree structure, we push the statistics :math:`g_i` and :math:`h_i` to the leaves they belong to,
|
||||
sum the statistics together, and use the formula to calculate how good the tree is.
|
||||
This score is like the impurity measure in a decision tree, except that it also takes the model complexity into account.
|
||||
|
||||
Learn the tree structure
|
||||
========================
|
||||
Now that we have a way to measure how good a tree is, ideally we would enumerate all possible trees and pick the best one.
|
||||
In practice this is intractable, so we will try to optimize one level of the tree at a time.
|
||||
Specifically we try to split a leaf into two leaves, and the score it gains is
|
||||
|
||||
.. math::
|
||||
Gain = \frac{1}{2} \left[\frac{G_L^2}{H_L+\lambda}+\frac{G_R^2}{H_R+\lambda}-\frac{(G_L+G_R)^2}{H_L+H_R+\lambda}\right] - \gamma
|
||||
|
||||
This formula can be decomposed as 1) the score on the new left leaf 2) the score on the new right leaf 3) The score on the original leaf 4) regularization on the additional leaf.
|
||||
We can see an important fact here: if the gain is smaller than :math:`\gamma`, we would do better not to add that branch. This is exactly the **pruning** techniques in tree based
|
||||
models! By using the principles of supervised learning, we can naturally come up with the reason these techniques work :)
|
||||
|
||||
For real valued data, we usually want to search for an optimal split. To efficiently do so, we place all the instances in sorted order, like the following picture.
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/model/split_find.png
|
||||
:width: 100%
|
||||
:alt: Schematic of choosing the best split
|
||||
|
||||
A left to right scan is sufficient to calculate the structure score of all possible split solutions, and we can find the best split efficiently.
|
||||
|
||||
**********************
|
||||
Final words on XGBoost
|
||||
**********************
|
||||
Now that you understand what boosted trees are, you may ask, where is the introduction for XGBoost?
|
||||
XGBoost is exactly a tool motivated by the formal principle introduced in this tutorial!
|
||||
More importantly, it is developed with both deep consideration in terms of **systems optimization** and **principles in machine learning**.
|
||||
The goal of this library is to push the extreme of the computation limits of machines to provide a **scalable**, **portable** and **accurate** library.
|
||||
Make sure you try it out, and most importantly, contribute your piece of wisdom (code, examples, tutorials) to the community!
|
||||
@ -1,90 +0,0 @@
|
||||
Monotonic Constraints
|
||||
=====================
|
||||
|
||||
It is often the case in a modeling problem or project that the functional form of an acceptable model is constrained in some way. This may happen due to business considerations, or because of the type of scientific question being investigated. In some cases, where there is a very strong prior belief that the true relationship has some quality, constraints can be used to improve the predictive performance of the model.
|
||||
|
||||
A common type of constraint in this situation is that certain features bear a *monotonic* relationship to the predicted response:
|
||||
|
||||
```math
|
||||
f(x_1, x_2, \ldots, x, \ldots, x_{n-1}, x_n) \leq f(x_1, x_2, \ldots, x', \ldots, x_{n-1}, x_n)
|
||||
```
|
||||
|
||||
whenever ``$ x \leq x' $`` is an *increasing constraint*; or
|
||||
|
||||
```math
|
||||
f(x_1, x_2, \ldots, x, \ldots, x_{n-1}, x_n) \geq f(x_1, x_2, \ldots, x', \ldots, x_{n-1}, x_n)
|
||||
```
|
||||
|
||||
whenever ``$ x \leq x' $`` is a *decreasing constraint*.
|
||||
|
||||
XGBoost has the ability to enforce monotonicity constraints on any features used in a boosted model.
|
||||
|
||||
A Simple Example
|
||||
----------------
|
||||
|
||||
To illustrate, let's create some simulated data with two features and a response according to the following scheme
|
||||
|
||||
```math
|
||||
y = 5 x_1 + \sin(10 \pi x_1) - 5 x_2 - \cos(10 \pi x_2) + N(0, 0.01)
|
||||
|
||||
x_1, x_2 \in [0, 1]
|
||||
```
|
||||
|
||||
The response generally increases with respect to the ``$ x_1 $`` feature, but a sinusoidal variation has been superimposed, resulting in the true effect being non-monotonic. For the ``$ x_2 $`` feature the variation is decreasing with a sinusoidal variation.
|
||||
|
||||
|
||||
|
||||
Let's fit a boosted tree model to this data without imposing any monotonic constraints
|
||||
|
||||
|
||||
|
||||
The black curve shows the trend inferred from the model for each feature. To make these plots the distinguished feature ``$x_i$`` is fed to the model over a one-dimensional grid of values, while all the other features (in this case only one other feature) are set to their average values. We see that the model does a good job of capturing the general trend with the oscillatory wave superimposed.
|
||||
|
||||
Here is the same model, but fit with monotonicity constraints
|
||||
|
||||
|
||||
|
||||
We see the effect of the constraint. For each variable the general direction of the trend is still evident, but the oscillatory behaviour no longer remains as it would violate our imposed constraints.
|
||||
|
||||
Enforcing Monotonic Constraints in XGBoost
|
||||
------------------------------------------
|
||||
|
||||
It is very simple to enforce monotonicity constraints in XGBoost. Here we will give an example using Python, but the same general idea generalizes to other platforms.
|
||||
|
||||
Suppose the following code fits your model without monotonicity constraints
|
||||
|
||||
```python
|
||||
model_no_constraints = xgb.train(params, dtrain,
|
||||
num_boost_round = 1000, evals = evallist,
|
||||
early_stopping_rounds = 10)
|
||||
```
|
||||
|
||||
Then fitting with monotonicity constraints only requires adding a single parameter
|
||||
|
||||
```python
|
||||
params_constrained = params.copy()
|
||||
params_constrained['monotone_constraints'] = "(1,-1)"
|
||||
|
||||
model_with_constraints = xgb.train(params_constrained, dtrain,
|
||||
num_boost_round = 1000, evals = evallist,
|
||||
early_stopping_rounds = 10)
|
||||
```
|
||||
|
||||
In this example the training data ```X``` has two columns, and by using the parameter values ```(1,-1)``` we are telling XGBoost to impose an increasing constraint on the first predictor and a decreasing constraint on the second.
|
||||
|
||||
Some other examples:
|
||||
|
||||
- ```(1,0)```: An increasing constraint on the first predictor and no constraint on the second.
|
||||
- ```(0,-1)```: No constraint on the first predictor and a decreasing constraint on the second.
|
||||
|
||||
**Choise of tree construction algorithm**. To use monotonic constraints, be
|
||||
sure to set the `tree_method` parameter to one of `'exact'`, `'hist'`, and
|
||||
`'gpu_hist'`.
|
||||
|
||||
**Note for the `'hist'` tree construction algorithm**.
|
||||
If `tree_method` is set to either `'hist'` or `'gpu_hist'`, enabling monotonic
|
||||
constraints may produce unnecessarily shallow trees. This is because the
|
||||
`'hist'` method reduces the number of candidate splits to be considered at each
|
||||
split. Monotonic constraints may wipe out all available split candidates, in
|
||||
which case no split is made. To reduce the effect, you may want to increase
|
||||
the `max_bin` parameter to consider more split candidates.
|
||||
95
doc/tutorials/monotonic.rst
Normal file
95
doc/tutorials/monotonic.rst
Normal file
@ -0,0 +1,95 @@
|
||||
#####################
|
||||
Monotonic Constraints
|
||||
#####################
|
||||
|
||||
It is often the case in a modeling problem or project that the functional form of an acceptable model is constrained in some way. This may happen due to business considerations, or because of the type of scientific question being investigated. In some cases, where there is a very strong prior belief that the true relationship has some quality, constraints can be used to improve the predictive performance of the model.
|
||||
|
||||
A common type of constraint in this situation is that certain features bear a **monotonic** relationship to the predicted response:
|
||||
|
||||
.. math::
|
||||
|
||||
f(x_1, x_2, \ldots, x, \ldots, x_{n-1}, x_n) \leq f(x_1, x_2, \ldots, x', \ldots, x_{n-1}, x_n)
|
||||
|
||||
whenever :math:`x \leq x'` is an **increasing constraint**; or
|
||||
|
||||
.. math::
|
||||
|
||||
f(x_1, x_2, \ldots, x, \ldots, x_{n-1}, x_n) \geq f(x_1, x_2, \ldots, x', \ldots, x_{n-1}, x_n)
|
||||
|
||||
whenever :math:`x \leq x'` is a **decreasing constraint**.
|
||||
|
||||
XGBoost has the ability to enforce monotonicity constraints on any features used in a boosted model.
|
||||
|
||||
****************
|
||||
A Simple Example
|
||||
****************
|
||||
|
||||
To illustrate, let's create some simulated data with two features and a response according to the following scheme
|
||||
|
||||
.. math::
|
||||
|
||||
y = 5 x_1 + \sin(10 \pi x_1) - 5 x_2 - \cos(10 \pi x_2) + N(0, 0.01)
|
||||
x_1, x_2 \in [0, 1]
|
||||
|
||||
The response generally increases with respect to the :math:`x_1` feature, but a sinusoidal variation has been superimposed, resulting in the true effect being non-monotonic. For the :math:`x_2` feature the variation is decreasing with a sinusoidal variation.
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/monotonic/two.feature.sample.data.png
|
||||
:alt: Data in sinusoidal fit
|
||||
|
||||
Let's fit a boosted tree model to this data without imposing any monotonic constraints:
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/monotonic/two.feature.no.constraint.png
|
||||
:alt: Fit of Model with No Constraint
|
||||
|
||||
The black curve shows the trend inferred from the model for each feature. To make these plots the distinguished feature :math:`x_i` is fed to the model over a one-dimensional grid of values, while all the other features (in this case only one other feature) are set to their average values. We see that the model does a good job of capturing the general trend with the oscillatory wave superimposed.
|
||||
|
||||
Here is the same model, but fit with monotonicity constraints:
|
||||
|
||||
.. image:: https://raw.githubusercontent.com/dmlc/web-data/master/xgboost/monotonic/two.feature.with.constraint.png
|
||||
:alt: Fit of Model with Constraint
|
||||
|
||||
We see the effect of the constraint. For each variable the general direction of the trend is still evident, but the oscillatory behaviour no longer remains as it would violate our imposed constraints.
|
||||
|
||||
******************************************
|
||||
Enforcing Monotonic Constraints in XGBoost
|
||||
******************************************
|
||||
|
||||
It is very simple to enforce monotonicity constraints in XGBoost. Here we will give an example using Python, but the same general idea generalizes to other platforms.
|
||||
|
||||
Suppose the following code fits your model without monotonicity constraints
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
model_no_constraints = xgb.train(params, dtrain,
|
||||
num_boost_round = 1000, evals = evallist,
|
||||
early_stopping_rounds = 10)
|
||||
|
||||
Then fitting with monotonicity constraints only requires adding a single parameter
|
||||
|
||||
.. code-block:: python
|
||||
|
||||
params_constrained = params.copy()
|
||||
params_constrained['monotone_constraints'] = "(1,-1)"
|
||||
|
||||
model_with_constraints = xgb.train(params_constrained, dtrain,
|
||||
num_boost_round = 1000, evals = evallist,
|
||||
early_stopping_rounds = 10)
|
||||
|
||||
In this example the training data ``X`` has two columns, and by using the parameter values ``(1,-1)`` we are telling XGBoost to impose an increasing constraint on the first predictor and a decreasing constraint on the second.
|
||||
|
||||
Some other examples:
|
||||
|
||||
- ``(1,0)``: An increasing constraint on the first predictor and no constraint on the second.
|
||||
- ``(0,-1)``: No constraint on the first predictor and a decreasing constraint on the second.
|
||||
|
||||
**Choise of tree construction algorithm**. To use monotonic constraints, be
|
||||
sure to set the ``tree_method`` parameter to one of ``exact``, ``hist``, and
|
||||
``gpu_hist``.
|
||||
|
||||
**Note for the 'hist' tree construction algorithm**.
|
||||
If ``tree_method`` is set to either ``hist`` or ``gpu_hist``, enabling monotonic
|
||||
constraints may produce unnecessarily shallow trees. This is because the
|
||||
``hist`` method reduces the number of candidate splits to be considered at each
|
||||
split. Monotonic constraints may wipe out all available split candidates, in
|
||||
which case no split is made. To reduce the effect, you may want to increase
|
||||
the ``max_bin`` parameter to consider more split candidates.
|
||||
@ -1,44 +1,55 @@
|
||||
#########################
|
||||
Notes on Parameter Tuning
|
||||
=========================
|
||||
#########################
|
||||
Parameter tuning is a dark art in machine learning, the optimal parameters
|
||||
of a model can depend on many scenarios. So it is impossible to create a
|
||||
comprehensive guide for doing so.
|
||||
|
||||
This document tries to provide some guideline for parameters in xgboost.
|
||||
|
||||
This document tries to provide some guideline for parameters in XGBoost.
|
||||
|
||||
************************************
|
||||
Understanding Bias-Variance Tradeoff
|
||||
------------------------------------
|
||||
************************************
|
||||
If you take a machine learning or statistics course, this is likely to be one
|
||||
of the most important concepts.
|
||||
When we allow the model to get more complicated (e.g. more depth), the model
|
||||
has better ability to fit the training data, resulting in a less biased model.
|
||||
However, such complicated model requires more data to fit.
|
||||
|
||||
Most of parameters in xgboost are about bias variance tradeoff. The best model
|
||||
Most of parameters in XGBoost are about bias variance tradeoff. The best model
|
||||
should trade the model complexity with its predictive power carefully.
|
||||
[Parameters Documentation](../parameter.md) will tell you whether each parameter
|
||||
will make the model more conservative or not. This can be used to help you
|
||||
:doc:`Parameters Documentation </parameter>` will tell you whether each parameter
|
||||
ill make the model more conservative or not. This can be used to help you
|
||||
turn the knob between complicated model and simple model.
|
||||
|
||||
*******************
|
||||
Control Overfitting
|
||||
-------------------
|
||||
When you observe high training accuracy, but low tests accuracy, it is likely that you encounter overfitting problem.
|
||||
*******************
|
||||
When you observe high training accuracy, but low test accuracy, it is likely that you encountered overfitting problem.
|
||||
|
||||
There are in general two ways that you can control overfitting in XGBoost
|
||||
|
||||
There are in general two ways that you can control overfitting in xgboost
|
||||
* The first way is to directly control model complexity
|
||||
- This include ```max_depth```, ```min_child_weight``` and ```gamma```
|
||||
* The second way is to add randomness to make training robust to noise
|
||||
- This include ```subsample```, ```colsample_bytree```
|
||||
- You can also reduce stepsize ```eta```, but needs to remember to increase ```num_round``` when you do so.
|
||||
|
||||
- This include ``max_depth``, ``min_child_weight`` and ``gamma``
|
||||
|
||||
* The second way is to add randomness to make training robust to noise
|
||||
|
||||
- This include ``subsample`` and ``colsample_bytree``.
|
||||
- You can also reduce stepsize ``eta``. Rremember to increase ``num_round`` when you do so.
|
||||
|
||||
*************************
|
||||
Handle Imbalanced Dataset
|
||||
-------------------------
|
||||
*************************
|
||||
For common cases such as ads clickthrough log, the dataset is extremely imbalanced.
|
||||
This can affect the training of xgboost model, and there are two ways to improve it.
|
||||
* If you care only about the ranking order (AUC) of your prediction
|
||||
- Balance the positive and negative weights, via ```scale_pos_weight```
|
||||
This can affect the training of XGBoost model, and there are two ways to improve it.
|
||||
|
||||
* If you care only about the overall performance metric (AUC) of your prediction
|
||||
|
||||
- Balance the positive and negative weights via ``scale_pos_weight``
|
||||
- Use AUC for evaluation
|
||||
|
||||
* If you care about predicting the right probability
|
||||
|
||||
- In such a case, you cannot re-balance the dataset
|
||||
- In such a case, set parameter ```max_delta_step``` to a finite number (say 1) will help convergence
|
||||
- Set parameter ``max_delta_step`` to a finite number (say 1) to help convergence
|
||||
Loading…
x
Reference in New Issue
Block a user